Volume 28, Issue 3 (Summer 2022)
Intern Med Today 2022, 28(3): 330-353 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Adeli H, Ahmadizadeh C, Sadeghi Zali M H. Investigation of the Effect of Lactobacillus Brevis Bacteria on the Expression of Rel A, IKB, and Casp3 Genes in HT29 Colon Cancer Cells. Intern Med Today 2022; 28 (3) :330-353
URL: http://imtj.gmu.ac.ir/article-1-3876-en.html
URL: http://imtj.gmu.ac.ir/article-1-3876-en.html
1- Department of Microbiology, Ahar Branch, Islamic Azad University, Ahar, Iran.
2- Department of Microbiology, Ahar Branch, Islamic Azad University, Ahar, Iran. ,dr_ahmadizadeh@yahoo.com
3- Department of Microbiology, Urmia branch, Islamic Azad University, Urmia, Iran.
2- Department of Microbiology, Ahar Branch, Islamic Azad University, Ahar, Iran. ,
3- Department of Microbiology, Urmia branch, Islamic Azad University, Urmia, Iran.
Full-Text [PDF 5671 kb]
(1102 Downloads)
| Abstract (HTML) (1611 Views)
Full-Text: (2688 Views)
Introduction
In Iran, colon cancer is one of the most common cancers related to the digestive system. Regarding suffering from this cancer, Iranian men rank third, and women rank fourth [1]. In vitro experiments showed that probiotics play a role in suppressing primary neoplastic ulcer and colon cancer tumors [2]. The anticancer effects of probiotics are manifested by preventing the transformation of procarcinogens into carcinogens, binding and inactivating mitogenic compounds, reducing the growth of procarcinogenic bacteria (carcinogenic agents), reducing the absorption of mitogens, and increasing the function of the immune system [3]. Also, one of the functional mechanisms of probiotics, including lactobacilli, is the anti-proliferative property of cancer cells, including colon cancer, by inducing apoptosis [4].
Colon cancer often develops in the form of polyps on the surface of the inner wall of the intestine, which originates from the inner lining of the large intestine. These masses are usually non-cancerous, but if left untreated, they may become colon cancer [5]. Various factors, such as genetic, environmental, and dietary factors, can be considered the cause of colon cancer [6, 7]. It has been found that pro-neoplastic factors are present in the colon of people with colon cancer. Evidence has shown that probiotics can play a role in the prevention and relief of colon cancer symptoms [8].
Apoptosis induction in cancer cells depends on the activation of PTEN/Akt and NF-kB signaling pathway subunits. The activation of the PTEN (tumor suppressor) gene in the PTEN/Akt signaling pathway and the Rel A gene in the NF-kB pathway inhibits the proliferation of colorectal cancer cells, induces apoptosis, and induces M/G2 cell cycle arrest [9, 10].
NF-KB pathway signaling (activation of Rel A, IKB subunits) is involved in cell proliferation, inflammation, and apoptosis [11]. The activation of TLR4 signaling leads to the activation of MAPK and AKT pathways and, finally, the activation of NF-KB (nuclear factor), transcription factors, and APL1 (activator protein) and control of the expression of pro-inflammatory cytokines and other genes related to immunity. TLR4 also activates IRF3 (interferon regulatory factor) and induces the expression of interferon (IFNB) and INF-responsive genes [12].
Probiotics are live and non-pathogenic microorganisms found in some foods that, when sufficient amounts of them enter the body, have a positive effect on the host’s health. Types of lactic acid bacteria including Lactobacillus species, Bifidobacterium species, Enterococcus faecium species, Lactococcus lactis, Leuconostoc mesantheroides, Pediococcus acid lactic, and Streptococcus thermophilus are probiotic microorganisms. Lactobacilli and Bifidobacterium are among the most commonly used bacteria as probiotics, but some yeasts and other bacilli are also used [13].
The belief in the beneficial effects of probiotics is based on the fact that intestinal microbial flora plays a protective role against various diseases. Probiotics’ main product is determined by stabilizing intestinal flora [14]. Parker defined probiotics as effective organisms that maintain intestinal microbial balance [15]. Probiotic means a product containing living and specific microorganisms with a sufficient number that colonization in a part of the body by creating a balance in the microbial flora and causing beneficial effects on the host’s health [16].
Wagner et al. stated that Lactobacillus roteri bacteria might prevent colorectal cancer by reducing the expression of nuclear factor of NF-κB (kappaB) dependent gene products [17]. The NF-KB transcription factor is one of the crucial pathways with fast action and the first response factor to harmful cellular stimuli [18]. We aim to investigate the effect of isolated Lactobacillus brevis bacteria on expressing growth-related genes Rel A, IKB, and casp3 in HT29 colon cancer cells.
Materials and Methods
This experimental-laboratory study was conducted at Tabriz Medical Science Microtechnology Research Center in 2019. L. b strain (ATCC 13648) was prepared as good bacteria (probiotics) in the Bacteria Collection Center of the Iran Scientific Research Organization. Also, the HT29 cell line was designed from Pasteur’s cell collection. Also, HT-29 human colon adenocarcinoma grade II cell line inside a flask containing 90 cc of RPMI1640 culture medium enriched with 10% Fetal Bovine Serum (FBS), 100 µL of antibiotics (Penicillin 0.1 μg/μL and streptomycin 0.1 μg/μL) was cultured in an incubator at 37°C and 5% carbon dioxide. Then the cells were passaged according to the calculations related to the desired cell concentration (seed density) to perform the experiments. The required amount of the cell suspension was brought to the desired volume by the complete culture medium and was investigated with a microscope after checking the flask; then, they were incubated at 37°C [19].
The cultivation of L. b bacteria was also carried out in a nutrient-liquid culture medium. A total of 68 grams of the prepared medium powder was dissolved in one liter of distilled water. An autoclave machine sterilized the medium at 121°C for 20 minutes. To prepare bacterial supernatant, L. b bacteria were cultured in MRS liquid culture medium (De Man, Rogosa, and Sharpe agar) and kept in an incubator at 37°C for optimal culture.
Microculture tetrazolium (MTT) test
Measuring cell proliferation and survival using a fluorescent marker
The cytotoxic effect of L. b bacteria on the mentioned cancer cells was investigated by colorimetric method using tetrazolium dye (chemical name 3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl Tetrazolium bromide) which is abbreviated as microculture tetrazolium (MTT). The MTT investigation aims to measure and reproduce using a fluorescent marker compared to the control group. First, an appropriate number of HT29 cells are cultured in each well (12,000 to 10,000 cells). After 24 h, the wells are controlled, and a proper amount of live thermophilus bacteria is added. Three different optical density (OD) types (0.5, 1, and 1.5) of bacteria are added.
The bar plates are incubated for 4 hours to affect the bacteria. The OD of bacteria is measured at a wavelength of 600 nm. The number of bacteria in OD1 is 8 x 109 cells per mL; for OD2, it is 1.6 x 109 cells per mL; for OD1.5, it is 1.2 x 109 cells per mL; for OD 5, it is 4 x 108 cells per mL. After each incubation, the well’s medium is discarded, and each well is replaced with 200 μL of fresh medium and 50 μL of MTT solution (2 mg/mL dissolved in phosphate buffer solution preparation [PBS]). Cells not treated with bacteria were used as control. Then the plates were incubated for another 4 h at 37°C in the dark. After that, the MTT solution was replaced with 200 µL of dimethyl sulfoxide (DMSO) along with 25 µL of Sorenson’s buffer (0.1 M glycine, 0.1 M sodium chloride (NaCl) with pH 10.5 optimized with 1 M sodium hydroxide [NaOH]). Then the plates were shaken for 15 minutes at 37°C.
To determine the percentage of cell viability (viability test), at this stage, the cells are stained with trypan blue in such a way that 100 µL of the solution containing the cells is taken from under the hood and poured into a 2cc tube, and 100 µL trypan blue was added and mixed well. Then, cell counting was done using Neobar slides. The obtained average was multiplied by 100, and the dilution factor in getting the number of cells in one millimeter of solution. Optical absorbance was measured at 570 nm with an ELISA reader. The percentage of cell survival in the negative control group was 100, and the rate of cells affected by different concentrations of bacteria was calculated by dividing the absorbance of the treated wells by the absorbance of the negative control multiplied by 100. A concentration of the tested compounds that reduces the percentage of cell life by half was considered an inhibitory concentration (IC50). This value was determined from the graph using Excel software [20].
4′,6-Diamidino-2-Phenylindole (DAPI) cell staining
The purpose of DAPI staining is the effect of bacteria on the degeneration of the nucleus of cancer cells. To directly investigate the impact of L. b on HT29 cells, after treating all cells with the conditioning medium for 24 hours, the supernatant of the cell culture medium was completely emptied. They were fixed by adding 60 µL of 4% paraformaldehyde to each well for 8 minutes. The cells inside each well were washed three times with 60 µL of PBS. Then PBS was kept inside the wells for 5 minutes each time. Then the cells inside each well were permeabilized with 60 µLof 0.1% Triton x-100 permeabilizing solution for 10 minutes, and then the cells were passaged. Then, 50 µL of DAPI dye was to the cells inside each well and waited for 10 minutes, and it was washed 3 times again with PBS for 5 minutes. Then, 50 µL of PBS was poured into each well, and the plates were placed in the refrigerator before taking pictures. Then the cells were evaluated with an Olympus IX81 inverted fluorescent microscope equipped with an Olympus DP70 camera (Olympus Corp., Tokyo, Japan) [21].
Extraction of ribonucleic acid (RNA) with Trizol
To extract ribonucleic acid (RNA), 500 µL of Trizol was poured into each well of a 6-well plate and placed at room temperature for 20 minutes until the cells were completely lysed. Then, the contents of each 6-well container were transferred to 2 cc tubes, and about 200 µL of chloroform was added to each tube. Then the tubes were centrifuged at 12,000 rpm for 10 minutes and transferred to new 2 cc tubes. Then, 2.5 times the sample volume of cold isopropanol was added to each tube and placed in a -70°C freezer for 24 h. Then the tubes were centrifuged at 12000 rpm for 10 minutes, and the liquid on the tubes was poured out. Then the tubes were dried, and 20 µL of diethyl pyrocarbonate (DEPC) was poured into each tube. Then OD and their concentration were measured in ng/mL with a nanodrop device. To check the quality of the extracted RNAs, 5 samples were randomly electrophoresed on a 2% agarose gel for 1 h at 80 V [22].
Complementary DNA (cDNA) synthesis
One μg of total RNA reverse transcribed with 0.2 μM universal hexamer primer. Then 1 µL (10 mM) of deoxynucleotide triphosphate (dNTP) and DEPC water were mixed and incubated at 65°C for 5 minutes. Then put it on ice and add 5 units of reverse transcriptase (RT) (Moloney murine leukemia virus [MMLV]) enzyme, 1x buffer for Moloney murine leukemia virus (MMLV) RT (one unit per µL of ribonuclease [RNase] inhibitor), and at the end, the total volume of each tube was 20 µL. Then the tubes were placed in the thermocycler machine, and the program was given for 10 minutes at 25°C, 60 minutes at 42°C, and 10 minutes at 72°C to synthesize cDNAs.
Extraction of genomic DNA of L. b bacteria
The culture medium containing bacteria was transferred to 2 cc tubes and centrifuged at 10000 rpm for 5 minutes. The supernatant of the tubes was poured out, and then 800 µl of lysis buffer containing sodium hydroxide and safety data sheet (SDS) was added to each tube. Then the tubes were placed in a Bain-Marie at 85°C for 30 minutes. In the next step, the tubes were placed in a freezer at -70°C for 10 minutes and then placed in a Bain-Marie at 85°C for 5-6 minutes. Then 700 µL of chloroform-isoamyl alcohol solution, containing 24 cc of chloroform and 1 cc of isoamyl alcohol, was added to each of the tubes, and in the next step, the tubes were centrifuged at 12000 rpm for 5 minutes. Then, the supernatant of the tubes containing DNA was transferred to new 2 cc tubes, then 1.5 times the volume of cold isopropanol was added to the new tubes, and they were placed in a freezer at -70°C overnight. Then, the samples were centrifuged at 12000 rpm for 5 minutes, the supernatant was thrown out, and after drying, 50 µL of DNAs-free deionized distilled water was added to each tube.
Design primer
The primers used were designed by Oligo5 software and BLAST by the NCBI website. Tekaposist synthesized the primers (Table 1).

Real-time PCR reaction (BIO-RAD iQ5, USA) was performed in 3 replicates and according to the temperature program (Table 2).
They were poured into special real-time PCR tubes, 1 µL of cDNA and 19 µL of Cybergreen Mastermix containing 1 µL of forward primer (0.2 μM), 1 µL of reverse primer (0.2 μM), 7 µL of DEPC and 10 µL of Mastermix 1x real-time. Then the tubes were placed in the real-time PCR machine, and the machine was run. The GAPDH gene was considered an internal control gene.
Statistical analysis
Regarding the results obtained for the expression level of genes, first, the obtained CTs for each gene were calculated by the 2-∆∆CT formula. Then, the average (repeated three times) of the obtained results was calculated by SPSS statistical software in each group. Then, the normal distribution of the results was checked by the Shapiro-Wilks test. A one-way analysis of variance (ANOVA) statistical test was used in data analysis. The least significant difference (LSD) test was used to compare the groups’ mean. The tests were considered significant when the P value was less than 0.05. Then the expression ratio of each gene compared to the reference gene was calculated.
Results
Lactobacillus lethality on HT29 cell line
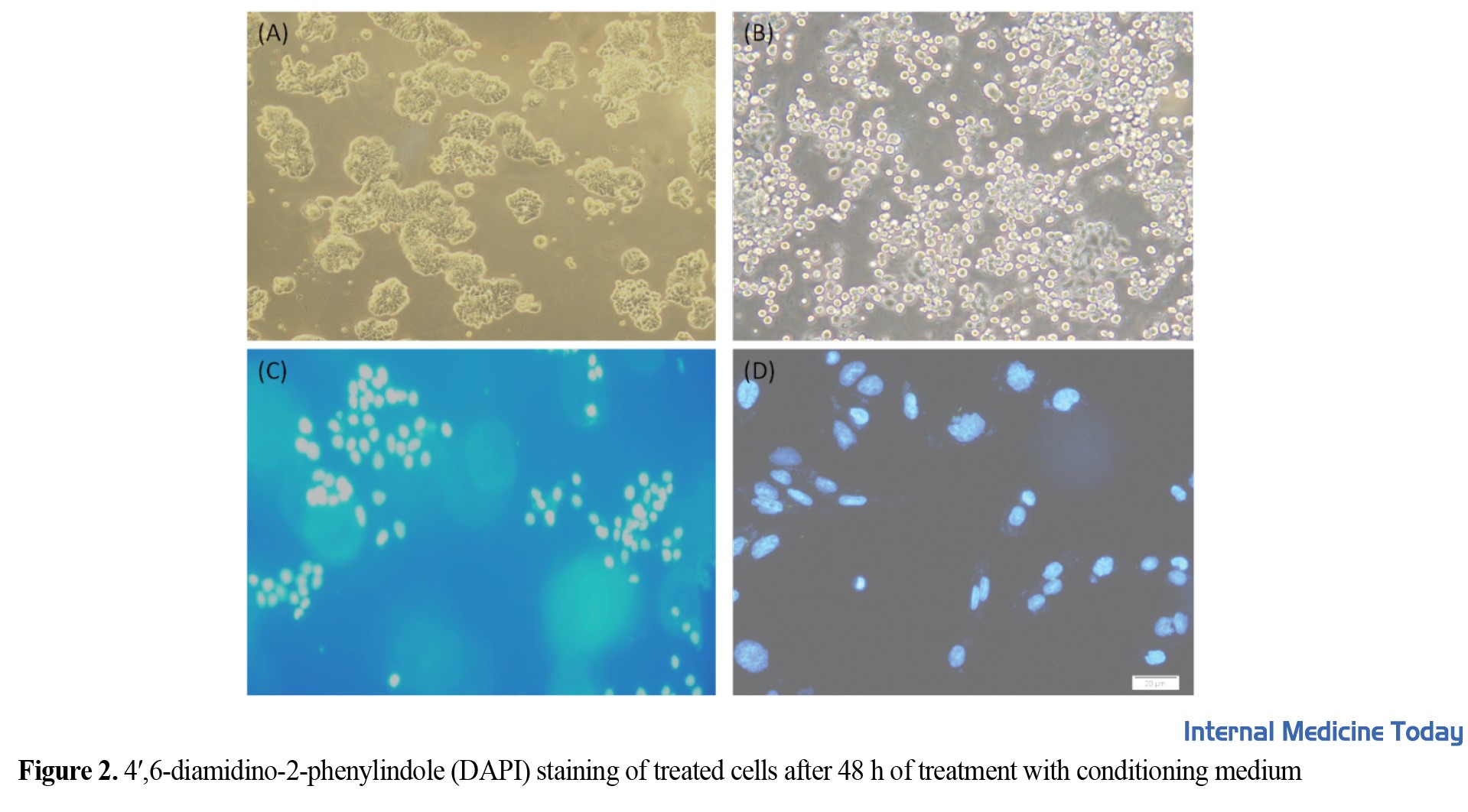
The number of cells required for the experiment was first optimized to investigate the lethal effect of L. b bacteria on HT29 cells. The mentioned cells were cultured in 10,000 cells per 96-well plate. Then these cells were incubated at 37°C. A day later, when the number of cells reached 20,000, i.e., 2 times, they were adjacent to increasing concentrations of bacteria. After 4 hours of being adjacent to the supernatant medium called conditioned media, they were filtered and added to new cells at different times. For each concentration of bacteria, 3 replications were considered, and 3 h were not treated with a conditioned environment as control. The results were obtained after 12, 24, and 48 h of treating the cells with a conditioned medium, L. b. Figure 1 and 2 shows the results of MTT. HT29 cells were affected by different concentrations of L. b bacteria. During 48 hours of incubation of HT29 cells with L. b bacteria, a specific IC50 value was obtained with a concentration of od=0.5. Also, after the cells’ treatment, DAPI staining was done on the cells to check the apoptosis. Figure 2 shows stained and unstained cells imaged by light and fluorescent microscopy. The treated cells entered apoptosis and formed smaller nuclei than untreated cells with a bacterial conditioning medium. A: Normal light microscopic healthy cells (HT29 cells grow normally). B: Healthy cells without normal light microscopy (HT29 cells undergo apoptosis or death under the influence of L. b bacteria). C: DAPI-stained cells under the fluorescent microscope and healthy without treatment with conditioned medium (shows the nucleus of HT29 cells, which are seen in blue with DAPI staining and without any change in a spherical shape). D: The nucleus of apoptotic cells stained with DAPI under a fluorescent microscope with fragmented nuclei (shows the nucleus of HT29 cells that are swollen and fragmented).
Polymerase chain reaction (PCR) for 16S rDNA gene of L. b bacteria
16S rDNA gene was used to confirm and determine the molecular identity of L. b by PCR. After PCR, the amplified fragment for the 16S rDNA gene is 1500 bp. After PCR, the product was electrophoresed in 2% agarose (Figure 3). A sharp band indicates specific amplification.
To perform this test, the cells were treated in 6-well plates with IC50 concentrations of the culture medium (condition) of L. b bacteria, and then the test was performed (Figure 4). It shows that along with the increase in the concentration of cDNAs, the samples with higher concentrations in lower cycles have reached the threshold. Since all factors except the number of cDNAs in the samples are constant, it is clear that the obtained CTs depends only on the concentration of cDNAs, which indicates the correctness of the work P=0.038.
Figures 2, 3, and 4 show the expression level of Rel A, IKB, and casp3 genes in the co-culture of HT29 colon cancer cells with L. b bacteria. They were treated individually for 12, 24, and 48 h and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant (***) (Figure 5). The expression of the Rel A gene in the HT29 cell line was evaluated after different dosages and throughout 12 to 48 h. The results showed that Rel A gene expression has a significant relationship in the first 48 h.
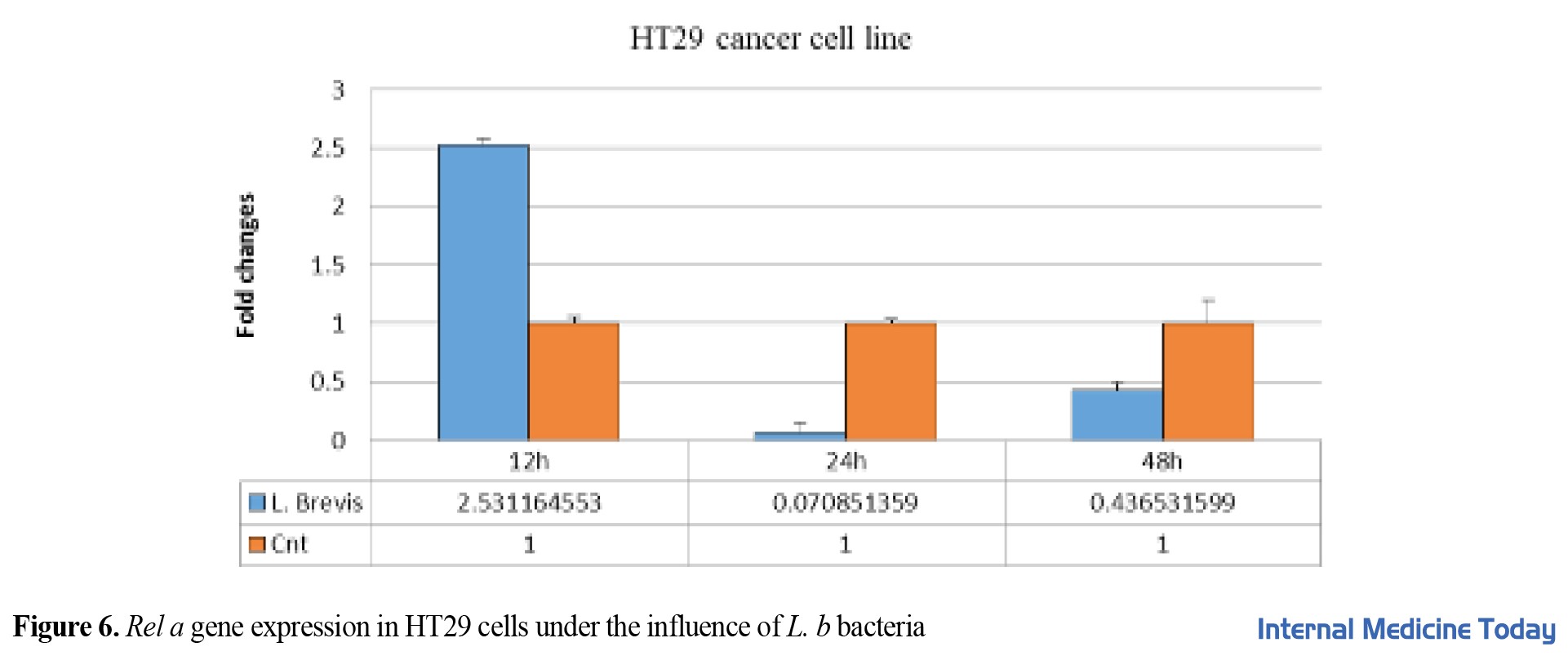
They were treated individually for 12, 24, and 48 h, and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant (***) P=0.038 (Figure 6). The expression of the IKB gene in the HT29 cell line was evaluated after different dosage proximity and in 12, 24, and 48 h. The results showed that the expression of the IKB gene has a significant relationship in the first 48 h P=0.042.
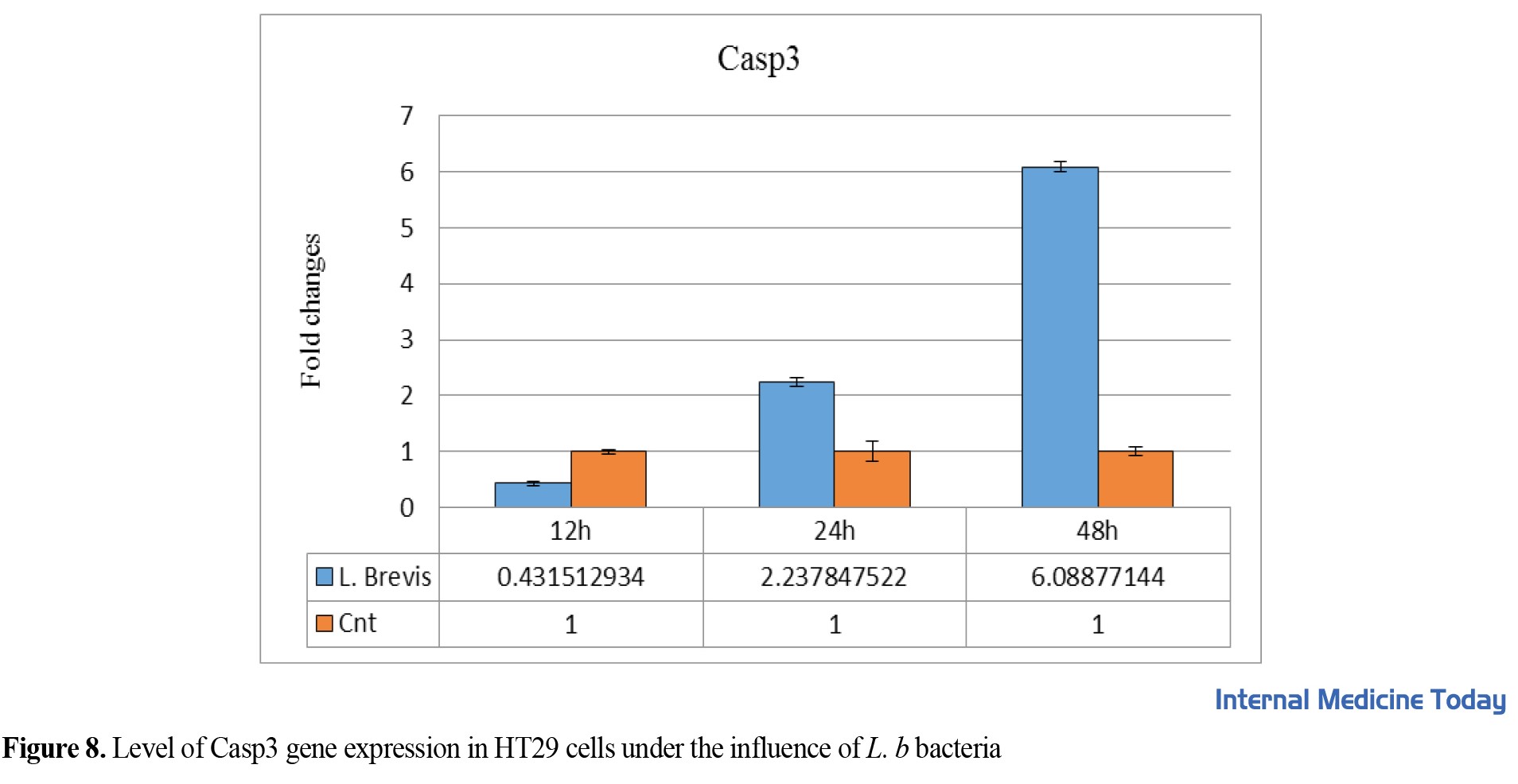
They were treated individually for 12, 24, and 48 h and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant (Figure 7). The expression of the casp3 gene in the HT29 cell line was evaluated after different dosages and for 12 to 24 h. The results showed that casp3 gene expression has a significant relationship in the first 48 h (P=0.038).
They were treated individually for 12, 24, and 48 h and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant, P=0.038 (Figure 7).
Discussion
Among all probiotics, Lactobacillus family bacteria, such as Lactobacillus acidophilus, Lactobacillus casei, and Lactobacillus delborki are among the vital components of the normal intestinal flora of humans and animals. In addition, the role of lactobacilli probiotics in facilitating the treatment of colorectal cancer is well known, which is consistent with our study [23, 24].
The current study evaluated the effect of L. b in preventing metastasis of colon cancer cells. The results of the MTT test showed that L. b inhibited the proliferation of HT29 cells and induced apoptosis in these cells, and inhibited the proliferation of the Rel A gene by increasing the expression of the IKB gene in these cells. The DAPI and DNA ladder assay results obtained from treating HT29 cells with the mentioned bacteria showed qualitative changes in cell apoptosis. In addition, real-time PCR results showed that L. b bacteria increased Casp3 gene expression in HT29 colon cancer cells.
According to the studies of al. Choi et al. showed that heat-killed Lactobacilli, including Lactobacillus acidophilus, L. b, and Lactobacillus casei, decreased the viability of cancer cell lines. In the current study, L. b decreased the viability of the HT-29 cell line; therefore, Choi et al.’s study is consistent with our study [25].
According to the studies conducted by Taverniti et al., in Lactobacillus plantarum and Lactobacillus casei, Lactobacillus bulgaricus decreased the viability of HT-29 and Caco-2 cells. It is consistent with our study [26].
Kim et al. suggested that compounds derived from probiotic bacteria can inhibit several cancers. Proteome analysis found that several proteins are involved in autophagy-mediated cell death, including GRP78 and Beclin-1, which are regulated by extracellular polysaccharides [27].
 In a study, Salva et al. showed that probiotic lactobacilli also protected against cyclophosphamide-induced mylo-phosphamide suppression in animal models, leading to improved resistance to Candida albicans. As a result, probiotics have been proposed as a way to reduce immunosuppression in cancer patients [28].
In a study, Salva et al. showed that probiotic lactobacilli also protected against cyclophosphamide-induced mylo-phosphamide suppression in animal models, leading to improved resistance to Candida albicans. As a result, probiotics have been proposed as a way to reduce immunosuppression in cancer patients [28].
Another study that confirms the current study was conducted by Sheng et al. They used Lactobacillus plantarum as a probiotic bacterium on HT29 cancer cells and introduced the PTEN pathway as the apoptosis pathway of cancer cells [29].
Chiu et al. also showed with their research that the soluble compounds secreted from Lactobacillus casei and Rhamnosus induce apoptosis in monocytic leukemia cells; as a result, probiotics can be considered a safe agent to fight cancer without any side effects, which is consistent with our study [30].
Liu & Pan also used ten native Lactobacillus strains in Taiwan and two lactic acid bacteria. They used cytoplasmic components and heat-killed bacteria on colon and breast cancer cell lines for their experiments. Their results showed that the inhibitory effect differs depending on the strain type, but the cells killed by heat decreased the viability percentage. In this study, L. b decreases the viability rate of the HT-29 cell line, which is consistent with our study [31].
Iyer et al. found that Lactobacillus roteri inhibited NF-κB induced by TNF activity in a dose- and time-dependent manner [32].
Pan et al. analyzed the effects of oral administration of Lactobacillus acidophilus bacteria on colorectal cancers in mice. The results indicated that Lactobacillus acidophilus reduced the severity of colorectal carcinogenesis and, on the other hand, increased programmed cell death in treated mice. In the current study, L. b also reduced colorectal carcinogens, which is consistent with our study [33].
In a study of clinical trials, Liu et al. showed the effect of probiotics in reducing infectious complications after surgery in colorectal cancer patients. Similarly, functional and quality-of-life health-related outcomes were significantly improved in patients undergoing resection for colorectal cancer treated with Lactobacillus acidophilus and Bacillus subtilis [34].
By investigating the effect of L. b on improving gastric gastritis caused by Helicobacter pylori infection in a mouse model, Orcid et al. showed that in infected groups, after treatment with L. b, inflammation was reduced and recovery was achieved. The rate of Helicobacter pylori infection eradicated in the treatment group showed a decrease in inflammation after tissue examination. In the current study, L. b reduces colorectal carcinogens and HT-29 cell line; therefore, the results of their study are consistent with our study [35].
Conclusion
Our findings show that L. b stimulates the cell signaling pathway of apoptosis in HT29 colon cancer cells and can be used as a new therapeutic strategy or adjuvant therapy for the treatment of colon cancer.
Ethical Considerations
Compliance with ethical guidelines
The Ethics Committee of the Ahar Branch, Islamic Azad University has approved this study (Code: 22030507971002).
Funding
This article is extracted from the MSc. thesis of Hojjat Adeli in the Department of Microbiology, Ahar Branch, Islamic Azad University.
Authors' contributions
All authors contributed equally in preparing all parts of the research.
Conflicts of interest
The authors declared no conflict of interest.
Acknowledgements
We express our gratitude to all those who helped us with this research.
References
In Iran, colon cancer is one of the most common cancers related to the digestive system. Regarding suffering from this cancer, Iranian men rank third, and women rank fourth [1]. In vitro experiments showed that probiotics play a role in suppressing primary neoplastic ulcer and colon cancer tumors [2]. The anticancer effects of probiotics are manifested by preventing the transformation of procarcinogens into carcinogens, binding and inactivating mitogenic compounds, reducing the growth of procarcinogenic bacteria (carcinogenic agents), reducing the absorption of mitogens, and increasing the function of the immune system [3]. Also, one of the functional mechanisms of probiotics, including lactobacilli, is the anti-proliferative property of cancer cells, including colon cancer, by inducing apoptosis [4].
Colon cancer often develops in the form of polyps on the surface of the inner wall of the intestine, which originates from the inner lining of the large intestine. These masses are usually non-cancerous, but if left untreated, they may become colon cancer [5]. Various factors, such as genetic, environmental, and dietary factors, can be considered the cause of colon cancer [6, 7]. It has been found that pro-neoplastic factors are present in the colon of people with colon cancer. Evidence has shown that probiotics can play a role in the prevention and relief of colon cancer symptoms [8].
Apoptosis induction in cancer cells depends on the activation of PTEN/Akt and NF-kB signaling pathway subunits. The activation of the PTEN (tumor suppressor) gene in the PTEN/Akt signaling pathway and the Rel A gene in the NF-kB pathway inhibits the proliferation of colorectal cancer cells, induces apoptosis, and induces M/G2 cell cycle arrest [9, 10].
NF-KB pathway signaling (activation of Rel A, IKB subunits) is involved in cell proliferation, inflammation, and apoptosis [11]. The activation of TLR4 signaling leads to the activation of MAPK and AKT pathways and, finally, the activation of NF-KB (nuclear factor), transcription factors, and APL1 (activator protein) and control of the expression of pro-inflammatory cytokines and other genes related to immunity. TLR4 also activates IRF3 (interferon regulatory factor) and induces the expression of interferon (IFNB) and INF-responsive genes [12].
Probiotics are live and non-pathogenic microorganisms found in some foods that, when sufficient amounts of them enter the body, have a positive effect on the host’s health. Types of lactic acid bacteria including Lactobacillus species, Bifidobacterium species, Enterococcus faecium species, Lactococcus lactis, Leuconostoc mesantheroides, Pediococcus acid lactic, and Streptococcus thermophilus are probiotic microorganisms. Lactobacilli and Bifidobacterium are among the most commonly used bacteria as probiotics, but some yeasts and other bacilli are also used [13].
The belief in the beneficial effects of probiotics is based on the fact that intestinal microbial flora plays a protective role against various diseases. Probiotics’ main product is determined by stabilizing intestinal flora [14]. Parker defined probiotics as effective organisms that maintain intestinal microbial balance [15]. Probiotic means a product containing living and specific microorganisms with a sufficient number that colonization in a part of the body by creating a balance in the microbial flora and causing beneficial effects on the host’s health [16].
Wagner et al. stated that Lactobacillus roteri bacteria might prevent colorectal cancer by reducing the expression of nuclear factor of NF-κB (kappaB) dependent gene products [17]. The NF-KB transcription factor is one of the crucial pathways with fast action and the first response factor to harmful cellular stimuli [18]. We aim to investigate the effect of isolated Lactobacillus brevis bacteria on expressing growth-related genes Rel A, IKB, and casp3 in HT29 colon cancer cells.
Materials and Methods
This experimental-laboratory study was conducted at Tabriz Medical Science Microtechnology Research Center in 2019. L. b strain (ATCC 13648) was prepared as good bacteria (probiotics) in the Bacteria Collection Center of the Iran Scientific Research Organization. Also, the HT29 cell line was designed from Pasteur’s cell collection. Also, HT-29 human colon adenocarcinoma grade II cell line inside a flask containing 90 cc of RPMI1640 culture medium enriched with 10% Fetal Bovine Serum (FBS), 100 µL of antibiotics (Penicillin 0.1 μg/μL and streptomycin 0.1 μg/μL) was cultured in an incubator at 37°C and 5% carbon dioxide. Then the cells were passaged according to the calculations related to the desired cell concentration (seed density) to perform the experiments. The required amount of the cell suspension was brought to the desired volume by the complete culture medium and was investigated with a microscope after checking the flask; then, they were incubated at 37°C [19].
The cultivation of L. b bacteria was also carried out in a nutrient-liquid culture medium. A total of 68 grams of the prepared medium powder was dissolved in one liter of distilled water. An autoclave machine sterilized the medium at 121°C for 20 minutes. To prepare bacterial supernatant, L. b bacteria were cultured in MRS liquid culture medium (De Man, Rogosa, and Sharpe agar) and kept in an incubator at 37°C for optimal culture.
Microculture tetrazolium (MTT) test
Measuring cell proliferation and survival using a fluorescent marker
The cytotoxic effect of L. b bacteria on the mentioned cancer cells was investigated by colorimetric method using tetrazolium dye (chemical name 3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl Tetrazolium bromide) which is abbreviated as microculture tetrazolium (MTT). The MTT investigation aims to measure and reproduce using a fluorescent marker compared to the control group. First, an appropriate number of HT29 cells are cultured in each well (12,000 to 10,000 cells). After 24 h, the wells are controlled, and a proper amount of live thermophilus bacteria is added. Three different optical density (OD) types (0.5, 1, and 1.5) of bacteria are added.
The bar plates are incubated for 4 hours to affect the bacteria. The OD of bacteria is measured at a wavelength of 600 nm. The number of bacteria in OD1 is 8 x 109 cells per mL; for OD2, it is 1.6 x 109 cells per mL; for OD1.5, it is 1.2 x 109 cells per mL; for OD 5, it is 4 x 108 cells per mL. After each incubation, the well’s medium is discarded, and each well is replaced with 200 μL of fresh medium and 50 μL of MTT solution (2 mg/mL dissolved in phosphate buffer solution preparation [PBS]). Cells not treated with bacteria were used as control. Then the plates were incubated for another 4 h at 37°C in the dark. After that, the MTT solution was replaced with 200 µL of dimethyl sulfoxide (DMSO) along with 25 µL of Sorenson’s buffer (0.1 M glycine, 0.1 M sodium chloride (NaCl) with pH 10.5 optimized with 1 M sodium hydroxide [NaOH]). Then the plates were shaken for 15 minutes at 37°C.
To determine the percentage of cell viability (viability test), at this stage, the cells are stained with trypan blue in such a way that 100 µL of the solution containing the cells is taken from under the hood and poured into a 2cc tube, and 100 µL trypan blue was added and mixed well. Then, cell counting was done using Neobar slides. The obtained average was multiplied by 100, and the dilution factor in getting the number of cells in one millimeter of solution. Optical absorbance was measured at 570 nm with an ELISA reader. The percentage of cell survival in the negative control group was 100, and the rate of cells affected by different concentrations of bacteria was calculated by dividing the absorbance of the treated wells by the absorbance of the negative control multiplied by 100. A concentration of the tested compounds that reduces the percentage of cell life by half was considered an inhibitory concentration (IC50). This value was determined from the graph using Excel software [20].
4′,6-Diamidino-2-Phenylindole (DAPI) cell staining
The purpose of DAPI staining is the effect of bacteria on the degeneration of the nucleus of cancer cells. To directly investigate the impact of L. b on HT29 cells, after treating all cells with the conditioning medium for 24 hours, the supernatant of the cell culture medium was completely emptied. They were fixed by adding 60 µL of 4% paraformaldehyde to each well for 8 minutes. The cells inside each well were washed three times with 60 µL of PBS. Then PBS was kept inside the wells for 5 minutes each time. Then the cells inside each well were permeabilized with 60 µLof 0.1% Triton x-100 permeabilizing solution for 10 minutes, and then the cells were passaged. Then, 50 µL of DAPI dye was to the cells inside each well and waited for 10 minutes, and it was washed 3 times again with PBS for 5 minutes. Then, 50 µL of PBS was poured into each well, and the plates were placed in the refrigerator before taking pictures. Then the cells were evaluated with an Olympus IX81 inverted fluorescent microscope equipped with an Olympus DP70 camera (Olympus Corp., Tokyo, Japan) [21].
Extraction of ribonucleic acid (RNA) with Trizol
To extract ribonucleic acid (RNA), 500 µL of Trizol was poured into each well of a 6-well plate and placed at room temperature for 20 minutes until the cells were completely lysed. Then, the contents of each 6-well container were transferred to 2 cc tubes, and about 200 µL of chloroform was added to each tube. Then the tubes were centrifuged at 12,000 rpm for 10 minutes and transferred to new 2 cc tubes. Then, 2.5 times the sample volume of cold isopropanol was added to each tube and placed in a -70°C freezer for 24 h. Then the tubes were centrifuged at 12000 rpm for 10 minutes, and the liquid on the tubes was poured out. Then the tubes were dried, and 20 µL of diethyl pyrocarbonate (DEPC) was poured into each tube. Then OD and their concentration were measured in ng/mL with a nanodrop device. To check the quality of the extracted RNAs, 5 samples were randomly electrophoresed on a 2% agarose gel for 1 h at 80 V [22].
Complementary DNA (cDNA) synthesis
One μg of total RNA reverse transcribed with 0.2 μM universal hexamer primer. Then 1 µL (10 mM) of deoxynucleotide triphosphate (dNTP) and DEPC water were mixed and incubated at 65°C for 5 minutes. Then put it on ice and add 5 units of reverse transcriptase (RT) (Moloney murine leukemia virus [MMLV]) enzyme, 1x buffer for Moloney murine leukemia virus (MMLV) RT (one unit per µL of ribonuclease [RNase] inhibitor), and at the end, the total volume of each tube was 20 µL. Then the tubes were placed in the thermocycler machine, and the program was given for 10 minutes at 25°C, 60 minutes at 42°C, and 10 minutes at 72°C to synthesize cDNAs.
Extraction of genomic DNA of L. b bacteria
The culture medium containing bacteria was transferred to 2 cc tubes and centrifuged at 10000 rpm for 5 minutes. The supernatant of the tubes was poured out, and then 800 µl of lysis buffer containing sodium hydroxide and safety data sheet (SDS) was added to each tube. Then the tubes were placed in a Bain-Marie at 85°C for 30 minutes. In the next step, the tubes were placed in a freezer at -70°C for 10 minutes and then placed in a Bain-Marie at 85°C for 5-6 minutes. Then 700 µL of chloroform-isoamyl alcohol solution, containing 24 cc of chloroform and 1 cc of isoamyl alcohol, was added to each of the tubes, and in the next step, the tubes were centrifuged at 12000 rpm for 5 minutes. Then, the supernatant of the tubes containing DNA was transferred to new 2 cc tubes, then 1.5 times the volume of cold isopropanol was added to the new tubes, and they were placed in a freezer at -70°C overnight. Then, the samples were centrifuged at 12000 rpm for 5 minutes, the supernatant was thrown out, and after drying, 50 µL of DNAs-free deionized distilled water was added to each tube.
Design primer
The primers used were designed by Oligo5 software and BLAST by the NCBI website. Tekaposist synthesized the primers (Table 1).

Real-time PCR reaction (BIO-RAD iQ5, USA) was performed in 3 replicates and according to the temperature program (Table 2).

They were poured into special real-time PCR tubes, 1 µL of cDNA and 19 µL of Cybergreen Mastermix containing 1 µL of forward primer (0.2 μM), 1 µL of reverse primer (0.2 μM), 7 µL of DEPC and 10 µL of Mastermix 1x real-time. Then the tubes were placed in the real-time PCR machine, and the machine was run. The GAPDH gene was considered an internal control gene.
Statistical analysis
Regarding the results obtained for the expression level of genes, first, the obtained CTs for each gene were calculated by the 2-∆∆CT formula. Then, the average (repeated three times) of the obtained results was calculated by SPSS statistical software in each group. Then, the normal distribution of the results was checked by the Shapiro-Wilks test. A one-way analysis of variance (ANOVA) statistical test was used in data analysis. The least significant difference (LSD) test was used to compare the groups’ mean. The tests were considered significant when the P value was less than 0.05. Then the expression ratio of each gene compared to the reference gene was calculated.
Results
Lactobacillus lethality on HT29 cell line
The number of cells required for the experiment was first optimized to investigate the lethal effect of L. b bacteria on HT29 cells. The mentioned cells were cultured in 10,000 cells per 96-well plate. Then these cells were incubated at 37°C. A day later, when the number of cells reached 20,000, i.e., 2 times, they were adjacent to increasing concentrations of bacteria. After 4 hours of being adjacent to the supernatant medium called conditioned media, they were filtered and added to new cells at different times. For each concentration of bacteria, 3 replications were considered, and 3 h were not treated with a conditioned environment as control. The results were obtained after 12, 24, and 48 h of treating the cells with a conditioned medium, L. b. Figure 1 and 2 shows the results of MTT. HT29 cells were affected by different concentrations of L. b bacteria. During 48 hours of incubation of HT29 cells with L. b bacteria, a specific IC50 value was obtained with a concentration of od=0.5. Also, after the cells’ treatment, DAPI staining was done on the cells to check the apoptosis. Figure 2 shows stained and unstained cells imaged by light and fluorescent microscopy. The treated cells entered apoptosis and formed smaller nuclei than untreated cells with a bacterial conditioning medium. A: Normal light microscopic healthy cells (HT29 cells grow normally). B: Healthy cells without normal light microscopy (HT29 cells undergo apoptosis or death under the influence of L. b bacteria). C: DAPI-stained cells under the fluorescent microscope and healthy without treatment with conditioned medium (shows the nucleus of HT29 cells, which are seen in blue with DAPI staining and without any change in a spherical shape). D: The nucleus of apoptotic cells stained with DAPI under a fluorescent microscope with fragmented nuclei (shows the nucleus of HT29 cells that are swollen and fragmented).
Polymerase chain reaction (PCR) for 16S rDNA gene of L. b bacteria
16S rDNA gene was used to confirm and determine the molecular identity of L. b by PCR. After PCR, the amplified fragment for the 16S rDNA gene is 1500 bp. After PCR, the product was electrophoresed in 2% agarose (Figure 3). A sharp band indicates specific amplification.
To perform this test, the cells were treated in 6-well plates with IC50 concentrations of the culture medium (condition) of L. b bacteria, and then the test was performed (Figure 4). It shows that along with the increase in the concentration of cDNAs, the samples with higher concentrations in lower cycles have reached the threshold. Since all factors except the number of cDNAs in the samples are constant, it is clear that the obtained CTs depends only on the concentration of cDNAs, which indicates the correctness of the work P=0.038.
Figures 2, 3, and 4 show the expression level of Rel A, IKB, and casp3 genes in the co-culture of HT29 colon cancer cells with L. b bacteria. They were treated individually for 12, 24, and 48 h and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant (***) (Figure 5). The expression of the Rel A gene in the HT29 cell line was evaluated after different dosages and throughout 12 to 48 h. The results showed that Rel A gene expression has a significant relationship in the first 48 h.
They were treated individually for 12, 24, and 48 h, and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant (***) P=0.038 (Figure 6). The expression of the IKB gene in the HT29 cell line was evaluated after different dosage proximity and in 12, 24, and 48 h. The results showed that the expression of the IKB gene has a significant relationship in the first 48 h P=0.042.
They were treated individually for 12, 24, and 48 h and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant (Figure 7). The expression of the casp3 gene in the HT29 cell line was evaluated after different dosages and for 12 to 24 h. The results showed that casp3 gene expression has a significant relationship in the first 48 h (P=0.038).
They were treated individually for 12, 24, and 48 h and the number of changes in HT29 gene expression was measured by real-time PCR. All experiments were repeated three times, and P<0.05 was considered significant, P=0.038 (Figure 7).
Discussion
Among all probiotics, Lactobacillus family bacteria, such as Lactobacillus acidophilus, Lactobacillus casei, and Lactobacillus delborki are among the vital components of the normal intestinal flora of humans and animals. In addition, the role of lactobacilli probiotics in facilitating the treatment of colorectal cancer is well known, which is consistent with our study [23, 24].
The current study evaluated the effect of L. b in preventing metastasis of colon cancer cells. The results of the MTT test showed that L. b inhibited the proliferation of HT29 cells and induced apoptosis in these cells, and inhibited the proliferation of the Rel A gene by increasing the expression of the IKB gene in these cells. The DAPI and DNA ladder assay results obtained from treating HT29 cells with the mentioned bacteria showed qualitative changes in cell apoptosis. In addition, real-time PCR results showed that L. b bacteria increased Casp3 gene expression in HT29 colon cancer cells.
According to the studies of al. Choi et al. showed that heat-killed Lactobacilli, including Lactobacillus acidophilus, L. b, and Lactobacillus casei, decreased the viability of cancer cell lines. In the current study, L. b decreased the viability of the HT-29 cell line; therefore, Choi et al.’s study is consistent with our study [25].
According to the studies conducted by Taverniti et al., in Lactobacillus plantarum and Lactobacillus casei, Lactobacillus bulgaricus decreased the viability of HT-29 and Caco-2 cells. It is consistent with our study [26].
Kim et al. suggested that compounds derived from probiotic bacteria can inhibit several cancers. Proteome analysis found that several proteins are involved in autophagy-mediated cell death, including GRP78 and Beclin-1, which are regulated by extracellular polysaccharides [27].

Another study that confirms the current study was conducted by Sheng et al. They used Lactobacillus plantarum as a probiotic bacterium on HT29 cancer cells and introduced the PTEN pathway as the apoptosis pathway of cancer cells [29].
Chiu et al. also showed with their research that the soluble compounds secreted from Lactobacillus casei and Rhamnosus induce apoptosis in monocytic leukemia cells; as a result, probiotics can be considered a safe agent to fight cancer without any side effects, which is consistent with our study [30].
Liu & Pan also used ten native Lactobacillus strains in Taiwan and two lactic acid bacteria. They used cytoplasmic components and heat-killed bacteria on colon and breast cancer cell lines for their experiments. Their results showed that the inhibitory effect differs depending on the strain type, but the cells killed by heat decreased the viability percentage. In this study, L. b decreases the viability rate of the HT-29 cell line, which is consistent with our study [31].
Iyer et al. found that Lactobacillus roteri inhibited NF-κB induced by TNF activity in a dose- and time-dependent manner [32].
Pan et al. analyzed the effects of oral administration of Lactobacillus acidophilus bacteria on colorectal cancers in mice. The results indicated that Lactobacillus acidophilus reduced the severity of colorectal carcinogenesis and, on the other hand, increased programmed cell death in treated mice. In the current study, L. b also reduced colorectal carcinogens, which is consistent with our study [33].
In a study of clinical trials, Liu et al. showed the effect of probiotics in reducing infectious complications after surgery in colorectal cancer patients. Similarly, functional and quality-of-life health-related outcomes were significantly improved in patients undergoing resection for colorectal cancer treated with Lactobacillus acidophilus and Bacillus subtilis [34].
By investigating the effect of L. b on improving gastric gastritis caused by Helicobacter pylori infection in a mouse model, Orcid et al. showed that in infected groups, after treatment with L. b, inflammation was reduced and recovery was achieved. The rate of Helicobacter pylori infection eradicated in the treatment group showed a decrease in inflammation after tissue examination. In the current study, L. b reduces colorectal carcinogens and HT-29 cell line; therefore, the results of their study are consistent with our study [35].
Conclusion
Our findings show that L. b stimulates the cell signaling pathway of apoptosis in HT29 colon cancer cells and can be used as a new therapeutic strategy or adjuvant therapy for the treatment of colon cancer.
Ethical Considerations
Compliance with ethical guidelines
The Ethics Committee of the Ahar Branch, Islamic Azad University has approved this study (Code: 22030507971002).
Funding
This article is extracted from the MSc. thesis of Hojjat Adeli in the Department of Microbiology, Ahar Branch, Islamic Azad University.
Authors' contributions
All authors contributed equally in preparing all parts of the research.
Conflicts of interest
The authors declared no conflict of interest.
Acknowledgements
We express our gratitude to all those who helped us with this research.
References
- Kich DM, Vincenzi A, Majolo F, Volkende Souza CF, Goettert MI. Probiotic: Effectiveness nutrition in cancer treatment and prevention. Nutricion Hospitalaria. 2016; 33(6):1430-7. [DOI:10.20960/nh.806] [PMID]
- Guandalini S, Cernat E, Moscoso D. Prebiotics and probiotics inirritable bowel syndromeand inflammatory bowel disease in children. Beneficial Microbes. 2015; 6(2):209-17. [DOI:10.3920/BM2014.0067] [PMID]
- Whelan K. Probiotics and prebiotics in the management of irritable bowel syndrome: A review of recent clinical trials and systematic reviews. Current Opinion in Clinical Nutrition and Metabolic Care. 2011; 14(6):581-7 [DOI:10.1097/MCO.0b013e32834b8082] [PMID]
- Barrons R, Tassone D. Use of Lactobacillus probiotics for bacterial genitourinary infections in women: A review. Clinical Therapeutics. 2008; 30(3):453-68. [DOI:10.1016/j.clinthera.2008.03.013] [PMID]
- Gertler R, Rosenberg R, Schuster T, Friess H. Defining a high-risk subgroup with colon cancer stages I and II for possible adjuvant therapy. European Journal of Cancer. 2009; 45(17):2992-9. [DOI:10.1016/j.ejca.2009.07.008] [PMID]
- Slattery ML, Curtin K, Anderson K, Ma KN, Edwards S, Leppert M, et al. Associations between dietary intake and Ki-ras mutations in colon tumors: A population-based study. Cancer Research. 2000; 60(24):6935-41. [PMID]
- Brink M, Weijenberg MP, de Goeij AF, SchoutenLJ, Koedijk FD, Roemen GM, et al. Fat and K-ras mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2004; 25(9):1619-28. [DOI:10.1093/carcin/bgh177] [PMID]
- Pretlow TP, Barrow BJ, Ashton WS, O’Riordan MA, Pretlow TG, Jurcisek JA, et al. Aberrant crypts: Putative preneoplastic foci in human colonic mucosa. Cancer Research. 1991; 51(5):1564-7. [PMID]
- Vasudevan KM, Gurumurthy S, Rangnekar VM. Suppression of PTEN expression by NF-kappa B prevents apoptosis. Molecular and Cellular Biology. 2004; 24(3):1007-21. [DOI:10.1128/MCB.24.3.1007-1021.2004] [PMID] [PMCID]
- Buchholz TA, Garg AK, Chakravarti N, Aggarwal BB, Esteva FJ, Kuerer HM, et al. The nuclear transcription factor kappaB/bcl-2 pathway corRel Ates with pathologic complete response to doxorubicin-based neoadjuvant chemotherapy in human breast cancer. Clinical Cancer Research. 2005; 11(23):8398-402. [DOI:10.1158/1078-0432.CCR-05-0885] [PMID]
- Rajan T, Benluvankar V, Vincent S. Saccharomyces cerevisiae-induced apoptosis of monolayer cervical cancer cells. Asian Journal of Pharmaceutical and Clinical Research. 2017; 10(8):63-66. [DOI:10.22159/ajpcr.2017.v10i8.18818]
- Sargent DJ, Goldberg RM, Jacobson SD, Macdonald JS, Labianca R, Haller DG, et al. A pooled analysis of adjuvant chemotherapy for resected colon cancer in elderly patients. The New England Journal of Medicine. 2001; 345(15):1091-7. [DOI:10.1056/NEJMoa010957] [PMID]
- Sevda ER, Kopara AT, Kivance M. Cytotoxic effects of various lactic acid bacteria on Caco-2 cells. Turkish Journal of Biology. 2015; 39(1):23-30. [DOI:10.3906/biy-1402-62]
- Salminen S, Bouley C, Bourtron-Ruault MC, Cummings JH, Franck A, Gibson GR, et al. Functional food science and gastrointestinal physiology and function. The British Journal of Nutrition. 1998; 80(S1):S147-71. [DOI:10.1079/BJN19980108] [PMID]
- Parker RB. Probiotics, the other half of the antibiotic story. Animal Nutrition & Health.1974; 29:4-8. [Link]
- Scherezenmeir J, de Verse M. Probiotics and synbiotics approaching a definition. The American Journal of Clinical Nutrition. 2001; 73(2): 361S-64. [DOI:10.1093/ajcn/73.2.361s]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nature Reviews. Cancer. 2009; 9(8):537-49. [DOI:10.1038/nrc2694] [PMID]
- Mumtaz PT, Bhat SA, Ahmad SM, Dar MA, Ahmed R, Urwat U, et al. LncRNAs and immunity: Watchdogs for host pathogen interactions. Biological Procedures Online. 2017; 19:3. [DOI:10.1186/s12575-017-0052-7] [PMID] [PMCID]
- Oliver MH, Harrison NK, Bishop JE, Cole PJ, Laurent GJ. A rapid and convenient assay for counting cells cultured in microwell plates: Application for assessment of growth factors. Journal of Cell Science. 1989; 92(Pt 3):513-8. [DOI:10.1242/jcs.92.3.513] [PMID]
- Javidnia K, Miri R, Amirghofran Z, Jafari A, Amoozegar Z . [Cytotoxic ityandanti microbial assessment of Euphoria hebecarpa (Persian)]. Iranian Journal of Pharmaceutical Research. 2004; 3(2):75-82. [Link]
- Enoki T, Yoshida Y, Lally J, Hatta H, Bonen A.Testosterone increases lactate transport, monocarboxylate transporter (MCT) 1 and MCT4 in ratskeletal muscle. The Journal of Physiology. 2006; 577(Pt 1):433-43. [DOI:10.1113/jphysiol.2006.115436] [PMID] [PMCID]
- Peterson SM, Freeman JL. RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. Journal of Visualized Experiments. 2009; (30):1470. [DOI:10.3791/1470]
- Daniluk U. Probiotics, the new approach for cancer prevention and/or potentialization of anti-cancer treatment. Journal of Clinical & Experimental Oncology. 2012; 1:2. [DOI:10.4172/2324-9110.1000e105]
- de Moreno de LeBlanc A, Matar C, LeBlanc N, Perdigón G. Effects of milk fermented by Lactobacillus helveticus R389 on a murine breast cancer model. Breast Cancer Research . 2005; 7(4):R477-86. [DOI:10.1186/bcr1032] [PMID] [PMCID]
- Choi SS, Kim Y, Han KS, You S, Oh S, Kim SH. Effects of Lactobacillus strains on cancer cell proliferation and oxidative stress in vitro. Letters in Applied Microbiology. 2006; 42(5):452-58. [DOI:10.1111/j.1472-765X.2006.01913.x] [PMID]
- Taverniti V, Guglielmetti S. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes & Nutrition. 2011; 6(3):261-74. [DOI:10.1007/s12263-011-0218-x] [PMID] [PMCID]
- Kim Y, Oh S, Yun HS, Oh S, Kim SH. Cell-bound exopolysaccharide from probiotic bacteria induces autophagic cell death of tumour cells. Letters in Applied Microbiology. 2010; 51(2):123-30. [DOI:10.1111/j.1472-765X.2010.02859.x] [PMID]
- Salva S, Marranzino G, Villena J, Agüero G, Alvarez S. Probiotic Lactobacillus strains protect against myelosuppression and immunosuppression in cyclophosphamide-treated mice. International Immunopharmacology. 2014; 22(1):209-21. [DOI:10.1016/j.intimp.2014.06.017] [PMID]
- Sheng H, Shao J, Morrow JD, Beauchamp RD, Dubois RN. Modulation of apoptosis andbcl-2 exprssion by prostaglandin E2 in human colon cancer cells. Cancer Research. 1998; 58(2):362-6. [PMID]
- Chiu YH, Hsieh YJ, Liao KW, Peng KC. Preferential promotion of apoptosis of monocytes by Lactobacillus casei rhamnosus soluble factors. Clinical Nutrition. 2010; 29(1):131-40. [DOI:10.1016/j.clnu.2009.07.004] [PMID]
- Liu C, Pan T. In vitro effects of lactic acid bacteria on cancer cell viability and antioxidant activity. Journal of Food and Drug Analysis. 2010; 18(2):77-86. [DOI:10.38212/2224-6614.2287]
- Iyer C, Kosters A, Sethi G, Kunnumakkara AB, Aggarwal BB, Versalovic J. Probiotic Lactobacillus reuteri promotes TNF induced apoptosis in human myeloid leukemia derived cells by modulation of NF-κB and MAPK signalling. Cellular Microbiology. 2008; 10(7):1442-52 [DOI:10.1111/j.1462-5822.2008.01137.x] [PMID]
- Pan X, Chen F, Wu T, Tang H, Zhao Z. The acid, bile tolerance and antimicrobial property of Lactobacillus acidophilus NIT. Food Control. 2009; 20:598-602. [DOI:10.1016/j.foodcont.2008.08.019]
- Liu Z, Qin H, Yang Z, Xia Y, Liu W, Yang J, et al. Randomised clinical trial the effects of perioperative probiotic treatment on barrier function and post operative infectious complications in colorectal cancer surgery a double blind study. Alimentary Pharmacology & Therapeutics. 2011; 33(1):50-63. [DOI:10.1111/j.1365-2036.2010.04492.x] [PMID]
- Asgari B, Kermanian F, Khalili F, Rohani Nojede Sadat Z, Yaslianifard S. [Influence of lactobacillus brevis on the recovery of gastric gastritis caused by Helicobacter pylori infection in a C57BL / 6 mouse model (Persian)]. Knowledge Health. 2018; 13(2):15-21. [Link]
Type of Study: Original |
Subject:
Basic Medical Science
Received: 2022/03/7 | Accepted: 2022/06/22 | Published: 2022/07/1
Received: 2022/03/7 | Accepted: 2022/06/22 | Published: 2022/07/1
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |