Volume 28, Issue 3 (Summer 2022)
Intern Med Today 2022, 28(3): 382-397 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Soheilian‑Khorzoghi M, Rostami‑Nejad M, Haddadi A, Yadegar A, Dabiri H. Comparison of the Lactobacillus and Bifidobacterium Population in Fecal Microbiome of Celiac Disease Patients on Gluten-Free Diet With Healthy Subjects. Intern Med Today 2022; 28 (3) :382-397
URL: http://imtj.gmu.ac.ir/article-1-3842-en.html
URL: http://imtj.gmu.ac.ir/article-1-3842-en.html
Mona Soheilian‑Khorzoghi1 

 , Mohammad Rostami‑Nejad2
, Mohammad Rostami‑Nejad2 

 , Azam Haddadi1
, Azam Haddadi1 

 , Abbas Yadegar3
, Abbas Yadegar3 

 , Hossein Dabiri *4
, Hossein Dabiri *4 




 , Mohammad Rostami‑Nejad2
, Mohammad Rostami‑Nejad2 

 , Azam Haddadi1
, Azam Haddadi1 

 , Abbas Yadegar3
, Abbas Yadegar3 

 , Hossein Dabiri *4
, Hossein Dabiri *4 


1- Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran.
2- Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran,Iran.
3- Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran,Iran.
4- Department of Microbiology, Faculty of Medical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran. ,hdabiri@sbmu.ac.ir
2- Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran,Iran.
3- Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran,Iran.
4- Department of Microbiology, Faculty of Medical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran. ,
Full-Text [PDF 4896 kb]
(774 Downloads)
| Abstract (HTML) (1593 Views)
Full-Text: (1273 Views)
Introduction
Celiac disease (CD) is a chronic autoimmune disease triggered by gluten and other environmental factors, such as intestinal microbiota in genetically predisposed persons. Gluten consumption is the main cause of clinical signs and symptoms of celiac disease. When susceptible people are exposed to gluten, the tissue transglutaminase enzyme leads to changes in this protein, and as a result, an inflammatory reaction is created due to the interaction between the immune system and the tissue of the small intestine, and this reaction in the intestinal mucosa causes atrophy of the intestinal villi, crypt hyperplasia, increased number of lymphocytes in the lamina propria, and malabsorption syndrome. Currently, eliminating gluten from the diet is the only available treatment for celiac disease [2]. A gluten-free diet leads to improvement in the clinical signs and symptoms of celiac disease, small intestinal mucosal damage, and intestinal epithelial integrity [3]. The prevalence of celiac disease is increasing every year and the basic mechanism of this disorder is not fully understood. Celiac disease usually appears in early childhood after exposure to gluten; however, the number of people with celiac disease who experience this disease in early and late adulthood is also increasing [4]. For this reason, other environmental factors can also play a role in the spread of this disease [5]. Among these environmental factors, we can mention the short duration of breastfeeding, intestinal infections, and changes in the digestive microbiota [6]. Gastrointestinal microbiota includes a large number of microbial species and the number of its genes is 150 times that of the host’s genome, and it has a vital role in human health and is involved in crucial functions of the host, such as body metabolism and physiology [7]. Few studies have been conducted regarding the role of digestive microbiota in celiac disease; however, the change in digestive microbiota-dysbiosis is a critical environmental factor in the pathogenesis of the celiac disease [8]. Digestive microbiota is vital in the maturation of immunity in the body and homeostasis in the intestine. The dysbiosis in the gastrointestinal microbiota may affect intestinal homeostasis and thus lead to an immune response to food antigens, such as gluten [9]. Most studies show that dysbiosis occurs in the digestive microbiota of people with celiac disease with active disease in the form of a significant decrease in the population of beneficial gram-positive bacteria, such as Bifidobacterium and Lactobacillus in duodenal and stool samples. This reduction provides suitable conditions for the colonization of pathogenic gram-negative bacteria in the mucosal surfaces of celiac patients [10]. Also, the results of studies conducted on a sample of twelve celiac patients show that the Bifidobacterium population has decreased in these patients [1]. The presence of the Bifidobacteria family in the digestive tract leads to beneficial effects on a person’s health, including creating resistance in the host against pathogens [11]. In addition, studies conducted in children with celiac disease show a decrease in the ratio of Lactobacillus and Bifidobacterium to Bacteroides [12]. Different strains of Lactobacillus species exert more inductive effects than suppressive effects on both the innate and acquired immune systems of the body. Lactobacillus casei strains lead to an increase in both local and systemic T cell-mediated responses to gluten [13]. Considering the importance of intestinal microbiota in the pathogenesis of celiac disease and the imbalance in the intestinal microbiota of people with this disease, the present study was conducted to investigate the balance of two beneficial microbes, Lactobacillus and Bifidobacterium, in the stool samples of patients with celiac disease compared to healthy individuals.
Materials and Methods
Study population and sampling
This is a case-control study from August 2018 to February 2018 on 20 people with the celiac disease under a gluten-free diet as a case group and 20 healthy people as a control group who were referred to the celiac clinic of the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences. This study was approved with the ethics code IR.SBMU.RIGLD.REC.1395.114 of the Ethics Committee of the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences and Health Services and with the registration code IRCT20171225038063N1 of the Iranian Clinical Trial Registration Center (IRCT). After obtaining approvals, all participants in this study were informed about the objectives of this research project. Before entering the study, all patients with celiac disease had antibodies against tissue transglutaminase (t-TG) and endomysial antibody (EMA) in their blood serum, and their disease diagnosed by histology according to Marsh classification (Marsh I-III) was also confirmed [14]. The inclusion criteria included having a gluten-free diet for at least 6 months before entering the study. Before the study, the consent form was completed and approved by each participant. The inclusion criteria included pregnant women with celiac disease, people with gastrointestinal diseases, such as Crohn’s ulcerative colitis, ulcerative colitis, or ulcerative colitis, people with short bowel syndrome, people who have consumed alcoholic beverages, or people addicted to illegal drugs, people with a history of mouth and stomach surgery, people with cancer or positive HIV, people with a history of taking steroid drugs 4 weeks before the study, antibiotics, and non-steroidal anti-inflammatory drugs, and people with clinically abnormal levels of urea, electrolytes, creatinine, or liver serum. Demographic information of the participants, including age, sex, and pathology result of the patients was prepared and recorded based on the Marsh classification as well as a questionnaire. Before the study (day zero) and during the study, stool samples from both patient and control groups were collected in special plastic containers and kept at -80°C until analysis.
Extraction and measurement of DNA concentration
Bacterial DNA was extracted from the stool samples of the study subjects by the FavorPrepTM Stool DNA Isolation Mini Kit (Favorgen® Biotech Corp., Pingtung, Taiwan) according to the instructions mentioned in this kit. Briefly, 200 mg of feces was placed in a sterile tube, containing 300 μL of SDE1 buffer and 20 μL of proteinase K (10 mg/mL), and the rest of the protocol was performed according to the kit manufacturer’s instructions. After DNA extraction, its purity and concentration were measured by measuring the absorbance ratio of A260/A280 nm by NanoDrop®ND-1000 spectrophotometer (Thermo Scientific, USA). If the absorption ratio of 260 to 280 is between 1.7 and 2, the DNA has sufficient purity to perform a polymerase chain reaction [15]. The extracted DNA was stored at -80°C. Two bacterial candidates, Lactobacillus and Bifidobacterium, were investigated.
Analysis of intestinal microbiota by real-time polymerase chain reaction (PCR) technique
The real-time polymerase chain reaction (PCR) technique was used to identify and measure the number of copies of the 16S rRNA gene related to Bifidobacterium species and to determine the number of this bacterium in the intestinal microbiota, specific Bifid-F primers with the oligonucleotide sequence -GGGATGCTGGTGTGGAAGAG-3’5 and Bifid-R with the oligonucleotide sequence 5’-TGCTCGCGTCCACTATCCAG-3’ was used. For Lactobacillus species, specific primers Lacto-F with the oligonucleotide sequence 5’-TGGATGCCTTGGCACTAG-3’ and Lacto-R with the oligonucleotide sequence 5´-AAATCTCCGGATCAAAGCTTAC-3´ was used. The number of reaction components, including 10 μL of BioFACT™ 2X Real-Time PCR Master Mix (For SYBR Green I, BIOFACT, South Korea) and 10 nM of each reverse primer and 2 μL of extracted DNA were calculated. Real-time PCR program for each replication as an initial denaturation temperature of 95°C for 15 min, 40 cycles with an initial denaturation at 95°C for 20 s, annealing of primers at 56°C for 30 s, elongation at 72°C for 20 s, followed by the melting curve step according to the device instructions was performed. After performing the polymerase chain reaction, the temperature range of 52°C to 95°C was considered to determine the melting temperature and consecutive readings with a temperature gradient (Ramp) of 0.5°C. Melting curve analysis was performed to confirm the specificity of amplification. Primer concentrations and thermocycler programs were optimized for each specific polymerase chain reaction. Standard curve to determine the number of copies of the Swedberg 16 ribosomal RNA gene of each of the candidate bacteria by producing a 10-fold dilution series from 101 to 1010 copies of the Swedberg 16 ribosomal RNA gene in each reaction using Escherichia coli strain DNA BL21 (Escherichia coli BL21 strain) was performed. The number of 16S rRNA gene copies in the group of candidate bacteria in stool samples was determined by comparing the cycling threshold (CT) values of the samples with standard curves. All reactions were performed in triplicate.
Statistical analysis was performed using SPSS statistical software version 21 (Armonk, NY: IBM Corp). Differences in demographic criteria between the study groups were evaluated using the Pearson chi-square test for categorical variables. Two different groups were compared by t test. The obtained results were expressed as mean ± standard deviation (SD). In all cases, the significance level of the tests is less than 0.05. All analyzes were assessed for gut microbiota abundance based on a threshold cycle number.
Results
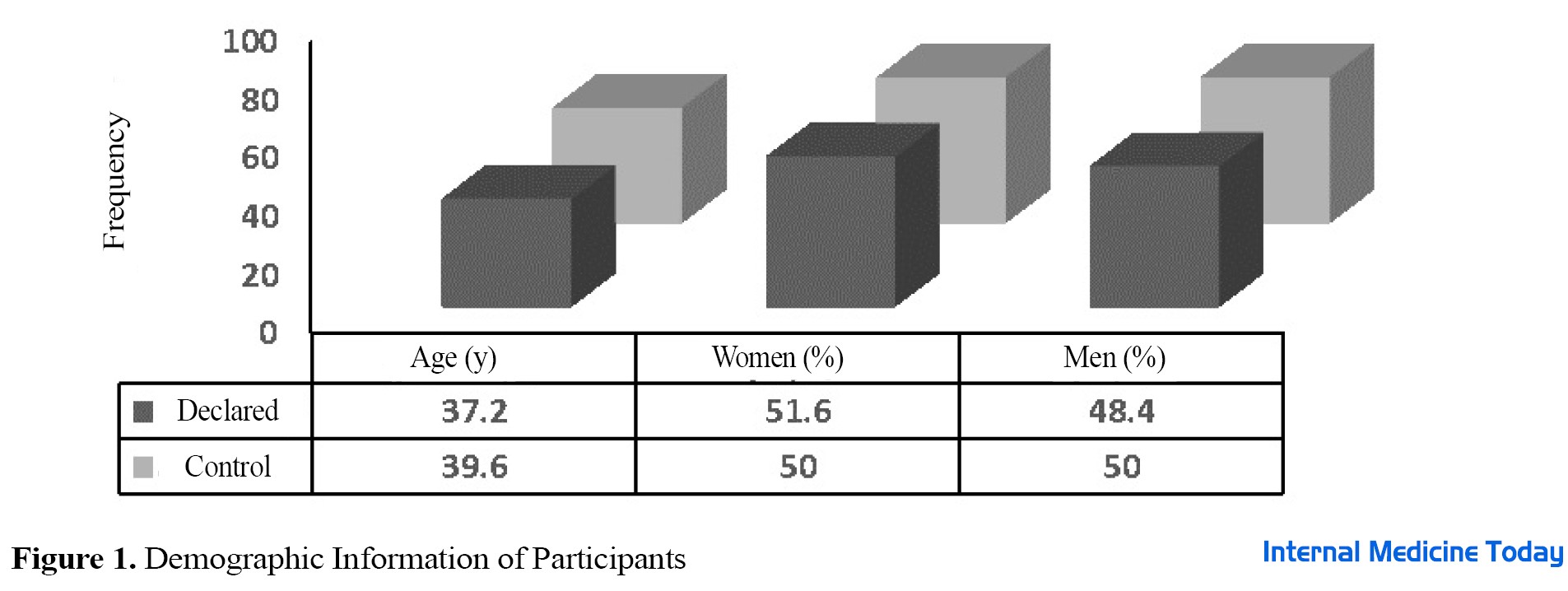
Demographic information in two groups of celiac patients under gluten-free and healthy diets was fully studied and statistically analyzed with related probability values (P value). Enumeration of intestinal microbiota in 20 people with celiac disease, including 9 men and 11 women, who were on a gluten-free diet (GFD) before entering the study and during the study as a case group, as well as 20 healthy people without celiac disease, including 10 men and 10 women, were examined as a control group in terms of composition. No statistically significant difference was observed in terms of gender and the average age of people in the two studied groups (P>0.05) (Figure 1). As shown in Figure 2, in terms of pathology results, most people with celiac disease were placed in Marsh group 3 (18 people). The results of the study showed no significant difference between the case group and control group in terms of Marsh classification (P>0.05). The number of target bacteria in fecal samples in both case and control groups was measured by the threshold cycle and shown as mean ± standard deviation in Table 1.
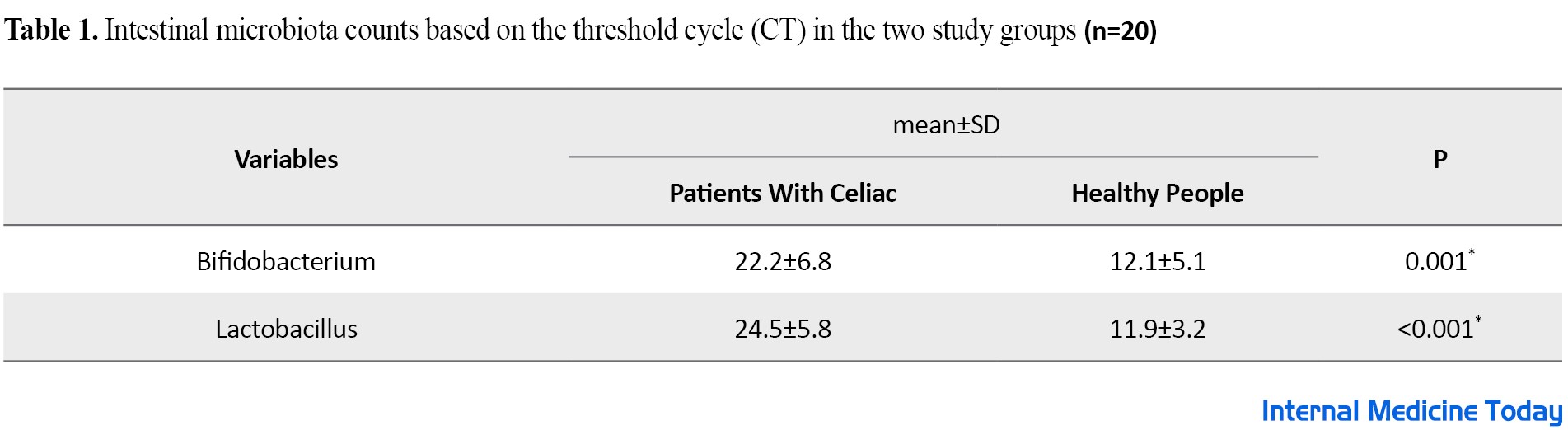
The results of this study show that the threshold cycle of Bifidobacterium and lactobacillus bacteria in people with celiac disease is significantly higher compared to the healthy group (P<0.05). Since the threshold cycle has an inverse relationship with the count rate, as a result, the count rate of these two useful bacteria in celiac patients is significantly lower compared to healthy people (P<0.05).
Discussion
Recent evidence regarding celiac disease has shown that innate immunity is vital in triggering the immune response through the stimulation of the acquired immune response and mucosal damage. The connection of intestinal microbiota with the intestinal mucosal wall is done through the same receptors that can activate innate immunity. Therefore, changes in gut microbiota may lead to the activation of this inflammatory pathway [16]. Beneficial species of the intestinal microbiota are reduced in patients with celiac disease, and on the other hand, pathogenic species are potentially increased compared to healthy people. In these patients, although dysbiosis in the intestinal microbiota is decreased after a GFD, it is not eliminated. Therefore, the intestinal microbiota plays a crucial role in the pathogenesis of the celiac disease [17, 18, 19]. On the other hand, fewer studies have investigated the number and composition of intestinal microbiota and its role in the pathogenesis of the celiac disease, as well as comparing the composition of intestinal microbiota in people with celiac disease compared to people without this disease. Therefore, in the current study, the composition of specific intestinal microbiota, including Bifidobacterium and Lactobacillus, was investigated in patients with celiac disease compared to healthy individuals, and it was shown that the population of intestinal microbiota in celiac patients is significantly different compared to healthy individuals so that sick people have a lower amount of beneficial intestinal bacteria Bifidobacterium and Lactobacillus than healthy people. The diversity of intestinal microbiota in people with celiac disease compared to healthy people has been investigated in various studies that confirm the results of the present study [10, 20]. Golfetto et al. agreed with the results of the present study regarding the low level of Bifidobacterium and dysbiosis in the intestinal microbiota in patients with celiac disease, even despite following a GFD, which supports the pathological process of the disease [21]. Similar to the present study is the study of Moraes et al. who reported the difference in the microbial profile between children with celiac disease and the control group and showed that celiac patients have a lower amount of Lactobacillus and Bifidobacterium compared to healthy individuals. Studies have shown that Bifidobacterium and Lactobacillus, which are also part of probiotics, can play a role in the digestion or change of gluten polypeptides. On the other hand, some bacterial species belonging to the genera Lactobacillus and Bifidobacterium have a protective role on epithelial cells against the damage caused by gliadin [22]. Collado et al. in a study identified specific intestinal bacteria related to celiac disease in the diagnosis and post-treatment with GFD in stool samples of children with untreated celiac disease and under GFD. Biopsy samples of untreated celiac patients under GFD and stool samples and biopsies of healthy children as a control group were examined to compare the two groups and real-time PCR was used to measure intestinal bacteria. The results of this study show the difference between the number of some intestinal microbiota, such as Bacteroidetes and Clostridium leptum in children with celiac disease compared to the control group regardless of the stage of the disease, as well as the difference in the number of E.coli. and showed Staphylococcus in untreated celiac children compared to the control group. Also, in this study, a lower amount of Bifidobacterium was reported in the feces of both groups of patients and biopsies of untreated individuals compared to the control group [10]. In another similar study, Sanz et al. analyzed the fecal microbiota count in the population of children with celiac disease compared to age-matched controls by a polymerase chain reaction and denaturing gradient gel electrophoresis and reported a significant increase in the stool microbiota population in patients with celiac disease compared to healthy individuals. On the other hand, they observed the specific presence of Lactobacillus curvatus species in celiac patients and Lactobacillus casei as a characteristic bacterial species in the healthy group. Also, the number of Bifidobacterium species in the celiac group was significantly lower than in the healthy group [21]. Nistal et al. conducted a study to investigate the difference in intestinal microbiota in adults with celiac disease and healthy individuals. Using the techniques of gradient genetic electrophoresis and gas-liquid chromatography of short-chain fatty acids, the microbial communities were measured in stool samples of untreated celiac patients, celiac patients treated with GFD, and healthy population and they observed a decrease in the diversity of Lactobacillus and Bifidobacterium species in treated celiac patients. Also, the treated celiac group showed a significantly higher amount of Bifidobacterium bifidum than healthy adults. The overall results of this study showed the difference in the fecal microbiota of untreated celiac patients compared to healthy people. Consistent with the present study, Nistal et al. showed that although the GFD in celiac patients partially restores the digestive microbiota to a normal state, the diversity of Lactobacillus and Bifidobacterium decreases significantly [23]. In another similar study conducted by Nylund et al. on patients with celiac disease and gluten-sensitive people without celiac disease under a GFD and healthy people consuming oats, the composition of fecal microbiota in the studied groups was investigated and the results showed that although the frequency of Bifidobacterium in healthy adults tended to be higher compared to celiac patients and the non-celiac gluten-sensitive group, it did not show a significant difference in terms of diversity in the microbial composition in the studied groups [33]. Also, Di Biase et al. compared the composition of the digestive microbiota in the stool samples of twelve children with celiac disease at the beginning of the disease with the healthy group. The results of this study are also similar to the current study, the difference in the digestive microbiota in children showed celiac disease at the beginning of the disease compared to the healthy group. A decrease in the abundance of beneficial bacteria, such as Bacteroides/Prevotella and Akkermansia was observed in stool samples of celiac disease patients compared to the healthy group [34]. In addition, some other studies also confirm the results of the present study and prove that two beneficial bacteria, Bifidobacterium and Lactobacillus, can play a protective role in patients with celiac disease against the inflammatory response and mucosal damage caused by gliadin peptides. They also explain the therapeutic roles of these probiotics [18, 19, 20, 21, 22, 23, 24]. Reducing the number of beneficial bacteria, such as Lactobacillus and Bifidobacterium in patients with celiac disease leads to an increase in opportunistic pathogens in patients with celiac disease and eventually leads to a defect in the immune system of these patients [26]. According to previous studies, the intestinal microbiota in patients with celiac disease is not completely restored despite following a GFD [10] and the amount of some useful digestive bacteria, such as Bifidobacterium in people with celiac disease is still significantly lower than in healthy people despite this type of diet. The results of most studies conducted in the field of investigating the diversity of the intestinal microbiota of celiac patients under a GFD and comparing it with healthy people all over the world show the diversity and a different number of the intestinal microbiota of these patients and healthy people. [27]. All these results confirm the results of the present study. In the current study, the target population was middle-aged patients and healthy people in an urban society with an average and normal diet. Compared to the group of healthy people, these people had an imbalance in the number of natural probiotics of flora. The amount of Lactobacillus and Bifidobacterium excreted in these people is less than in the healthy group, which is a reflection of the condition of the intestinal flora of these people, which is caused by the poor condition of the microbial flora and the lack of probiotic bacteria in the digestive system of these people. On the other hand, some studies showed no difference between the intestinal microbiota of patients with the celiac disease under a GFD and the control group and declared that the intestinal microbiota does not seem to play a role in the pathogenesis of the celiac disease [31]. On the other hand, having this type of diet in these patients, based on the results of previous studies, only leads to the improvement of a part of the intestinal microbiota [32], but the reasons are not known. However, factors such as the genetics of a patient with celiac disease, despite a GFD, can most likely affect the composition of the intestinal microbiota [33]. On the other hand, gluten has a prebiotic-like function, and removing this protein from a GFD can also create a different intestinal microbiota in these patients compared to healthy people [34]. Therefore, it can be said that modifying the nutritional system with the approach of strengthening the probiotic system of the digestive system and regulating their reliability and viability can moderate the complications of celiac disease, which is a kind of defect in the gene system with epigenetic stimuli. Physiologically, according to the atrophy of the intestinal villi in this disease, perhaps the defect in maintaining the balance of the natural microbial flora can be considered as one of the complications caused by this disease, in another approach, this factor can be considered as the cause of the exacerbation of this disease. According to other effective environmental and biological factors, this series of factors is the occurrence of nutritional poverty and deficiencies in the natural levels of micronutrients. Factors in the present study, the intestinal microbiota and the decrease in the amount of Bifidobacterium and Lactobacillus were different between the two studied groups and decreased in the group with celiac disease, but according to the sample size reported, this difference between the microbiota cannot be determined with certainty. Rohedei reported that the studies on healthy and celiac patients need to have a larger sample size. As shown in the findings section, the results of two Pearson chi-square tests for categorical variables and t test for numerical variables, which were performed to determine the integrity between the two groups of cases and controls in the statistical analysis of demographic data, show that the two studied groups have no significant differences in terms of demographic data. This problem is the strength of the present study in the sense that these results have confirmed the integration between the two studied groups and the two groups can be easily compared and examined. The present study had limitations. First, this study was conducted in a small sample size and a single center in Tehran City, Iran. Another limitation of the present study was the limited duration of the study and patient selection.
Conclusion
The present study, consistent with other studies, shows the phenomenon of imbalance of intestinal microbiota in patients with celiac disease compared to healthy people. As a result, it can be said that examining stool microbiota can be a good indicator of intestinal microbiota imbalance in celiac patients and can be used to monitor microbiota restoration during a gluten-free diet. The metabolic data along with the assessment of the intestinal microbiota of patients can help doctors evaluate the role of intestinal microbiota in the pathogenesis of celiac disease and also further investigations.
Ethical Considerations
Compliance with ethical guidelines
Following the principles of research ethics, the researchers followed all ethical codes related to research on human samples and obtained the necessary permits from the competent authorities from the Ethics Committee of the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences and Health Services (Ethical Code: IR.SBMU.RIGLD.REC.1395.114). Also, registration code IRCT20171225038063N1 of the Iran Clinical Trial Registration Center (IRCT) was obtained.
Funding
This research received financial support from the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences and Health Services.
Authors' contributions
All authors equally contributed to preparing this article.
Conflicts of interest
The authors declared no conflict of interest.
Acknowledgements
The authors of this article are grateful for the unwavering support of the vice president of research and the colleagues of the Research Institute of Gastrointestinal and Liver Diseases of Shahid Beheshti University of Medical Sciences and the respected officials of the Islamic Azad University, Karaj branch.
References
Celiac disease (CD) is a chronic autoimmune disease triggered by gluten and other environmental factors, such as intestinal microbiota in genetically predisposed persons. Gluten consumption is the main cause of clinical signs and symptoms of celiac disease. When susceptible people are exposed to gluten, the tissue transglutaminase enzyme leads to changes in this protein, and as a result, an inflammatory reaction is created due to the interaction between the immune system and the tissue of the small intestine, and this reaction in the intestinal mucosa causes atrophy of the intestinal villi, crypt hyperplasia, increased number of lymphocytes in the lamina propria, and malabsorption syndrome. Currently, eliminating gluten from the diet is the only available treatment for celiac disease [2]. A gluten-free diet leads to improvement in the clinical signs and symptoms of celiac disease, small intestinal mucosal damage, and intestinal epithelial integrity [3]. The prevalence of celiac disease is increasing every year and the basic mechanism of this disorder is not fully understood. Celiac disease usually appears in early childhood after exposure to gluten; however, the number of people with celiac disease who experience this disease in early and late adulthood is also increasing [4]. For this reason, other environmental factors can also play a role in the spread of this disease [5]. Among these environmental factors, we can mention the short duration of breastfeeding, intestinal infections, and changes in the digestive microbiota [6]. Gastrointestinal microbiota includes a large number of microbial species and the number of its genes is 150 times that of the host’s genome, and it has a vital role in human health and is involved in crucial functions of the host, such as body metabolism and physiology [7]. Few studies have been conducted regarding the role of digestive microbiota in celiac disease; however, the change in digestive microbiota-dysbiosis is a critical environmental factor in the pathogenesis of the celiac disease [8]. Digestive microbiota is vital in the maturation of immunity in the body and homeostasis in the intestine. The dysbiosis in the gastrointestinal microbiota may affect intestinal homeostasis and thus lead to an immune response to food antigens, such as gluten [9]. Most studies show that dysbiosis occurs in the digestive microbiota of people with celiac disease with active disease in the form of a significant decrease in the population of beneficial gram-positive bacteria, such as Bifidobacterium and Lactobacillus in duodenal and stool samples. This reduction provides suitable conditions for the colonization of pathogenic gram-negative bacteria in the mucosal surfaces of celiac patients [10]. Also, the results of studies conducted on a sample of twelve celiac patients show that the Bifidobacterium population has decreased in these patients [1]. The presence of the Bifidobacteria family in the digestive tract leads to beneficial effects on a person’s health, including creating resistance in the host against pathogens [11]. In addition, studies conducted in children with celiac disease show a decrease in the ratio of Lactobacillus and Bifidobacterium to Bacteroides [12]. Different strains of Lactobacillus species exert more inductive effects than suppressive effects on both the innate and acquired immune systems of the body. Lactobacillus casei strains lead to an increase in both local and systemic T cell-mediated responses to gluten [13]. Considering the importance of intestinal microbiota in the pathogenesis of celiac disease and the imbalance in the intestinal microbiota of people with this disease, the present study was conducted to investigate the balance of two beneficial microbes, Lactobacillus and Bifidobacterium, in the stool samples of patients with celiac disease compared to healthy individuals.
Materials and Methods
Study population and sampling
This is a case-control study from August 2018 to February 2018 on 20 people with the celiac disease under a gluten-free diet as a case group and 20 healthy people as a control group who were referred to the celiac clinic of the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences. This study was approved with the ethics code IR.SBMU.RIGLD.REC.1395.114 of the Ethics Committee of the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences and Health Services and with the registration code IRCT20171225038063N1 of the Iranian Clinical Trial Registration Center (IRCT). After obtaining approvals, all participants in this study were informed about the objectives of this research project. Before entering the study, all patients with celiac disease had antibodies against tissue transglutaminase (t-TG) and endomysial antibody (EMA) in their blood serum, and their disease diagnosed by histology according to Marsh classification (Marsh I-III) was also confirmed [14]. The inclusion criteria included having a gluten-free diet for at least 6 months before entering the study. Before the study, the consent form was completed and approved by each participant. The inclusion criteria included pregnant women with celiac disease, people with gastrointestinal diseases, such as Crohn’s ulcerative colitis, ulcerative colitis, or ulcerative colitis, people with short bowel syndrome, people who have consumed alcoholic beverages, or people addicted to illegal drugs, people with a history of mouth and stomach surgery, people with cancer or positive HIV, people with a history of taking steroid drugs 4 weeks before the study, antibiotics, and non-steroidal anti-inflammatory drugs, and people with clinically abnormal levels of urea, electrolytes, creatinine, or liver serum. Demographic information of the participants, including age, sex, and pathology result of the patients was prepared and recorded based on the Marsh classification as well as a questionnaire. Before the study (day zero) and during the study, stool samples from both patient and control groups were collected in special plastic containers and kept at -80°C until analysis.
Extraction and measurement of DNA concentration
Bacterial DNA was extracted from the stool samples of the study subjects by the FavorPrepTM Stool DNA Isolation Mini Kit (Favorgen® Biotech Corp., Pingtung, Taiwan) according to the instructions mentioned in this kit. Briefly, 200 mg of feces was placed in a sterile tube, containing 300 μL of SDE1 buffer and 20 μL of proteinase K (10 mg/mL), and the rest of the protocol was performed according to the kit manufacturer’s instructions. After DNA extraction, its purity and concentration were measured by measuring the absorbance ratio of A260/A280 nm by NanoDrop®ND-1000 spectrophotometer (Thermo Scientific, USA). If the absorption ratio of 260 to 280 is between 1.7 and 2, the DNA has sufficient purity to perform a polymerase chain reaction [15]. The extracted DNA was stored at -80°C. Two bacterial candidates, Lactobacillus and Bifidobacterium, were investigated.
Analysis of intestinal microbiota by real-time polymerase chain reaction (PCR) technique
The real-time polymerase chain reaction (PCR) technique was used to identify and measure the number of copies of the 16S rRNA gene related to Bifidobacterium species and to determine the number of this bacterium in the intestinal microbiota, specific Bifid-F primers with the oligonucleotide sequence -GGGATGCTGGTGTGGAAGAG-3’5 and Bifid-R with the oligonucleotide sequence 5’-TGCTCGCGTCCACTATCCAG-3’ was used. For Lactobacillus species, specific primers Lacto-F with the oligonucleotide sequence 5’-TGGATGCCTTGGCACTAG-3’ and Lacto-R with the oligonucleotide sequence 5´-AAATCTCCGGATCAAAGCTTAC-3´ was used. The number of reaction components, including 10 μL of BioFACT™ 2X Real-Time PCR Master Mix (For SYBR Green I, BIOFACT, South Korea) and 10 nM of each reverse primer and 2 μL of extracted DNA were calculated. Real-time PCR program for each replication as an initial denaturation temperature of 95°C for 15 min, 40 cycles with an initial denaturation at 95°C for 20 s, annealing of primers at 56°C for 30 s, elongation at 72°C for 20 s, followed by the melting curve step according to the device instructions was performed. After performing the polymerase chain reaction, the temperature range of 52°C to 95°C was considered to determine the melting temperature and consecutive readings with a temperature gradient (Ramp) of 0.5°C. Melting curve analysis was performed to confirm the specificity of amplification. Primer concentrations and thermocycler programs were optimized for each specific polymerase chain reaction. Standard curve to determine the number of copies of the Swedberg 16 ribosomal RNA gene of each of the candidate bacteria by producing a 10-fold dilution series from 101 to 1010 copies of the Swedberg 16 ribosomal RNA gene in each reaction using Escherichia coli strain DNA BL21 (Escherichia coli BL21 strain) was performed. The number of 16S rRNA gene copies in the group of candidate bacteria in stool samples was determined by comparing the cycling threshold (CT) values of the samples with standard curves. All reactions were performed in triplicate.
Statistical analysis was performed using SPSS statistical software version 21 (Armonk, NY: IBM Corp). Differences in demographic criteria between the study groups were evaluated using the Pearson chi-square test for categorical variables. Two different groups were compared by t test. The obtained results were expressed as mean ± standard deviation (SD). In all cases, the significance level of the tests is less than 0.05. All analyzes were assessed for gut microbiota abundance based on a threshold cycle number.
Results
Demographic information in two groups of celiac patients under gluten-free and healthy diets was fully studied and statistically analyzed with related probability values (P value). Enumeration of intestinal microbiota in 20 people with celiac disease, including 9 men and 11 women, who were on a gluten-free diet (GFD) before entering the study and during the study as a case group, as well as 20 healthy people without celiac disease, including 10 men and 10 women, were examined as a control group in terms of composition. No statistically significant difference was observed in terms of gender and the average age of people in the two studied groups (P>0.05) (Figure 1). As shown in Figure 2, in terms of pathology results, most people with celiac disease were placed in Marsh group 3 (18 people). The results of the study showed no significant difference between the case group and control group in terms of Marsh classification (P>0.05). The number of target bacteria in fecal samples in both case and control groups was measured by the threshold cycle and shown as mean ± standard deviation in Table 1.

The results of this study show that the threshold cycle of Bifidobacterium and lactobacillus bacteria in people with celiac disease is significantly higher compared to the healthy group (P<0.05). Since the threshold cycle has an inverse relationship with the count rate, as a result, the count rate of these two useful bacteria in celiac patients is significantly lower compared to healthy people (P<0.05).
Discussion
Recent evidence regarding celiac disease has shown that innate immunity is vital in triggering the immune response through the stimulation of the acquired immune response and mucosal damage. The connection of intestinal microbiota with the intestinal mucosal wall is done through the same receptors that can activate innate immunity. Therefore, changes in gut microbiota may lead to the activation of this inflammatory pathway [16]. Beneficial species of the intestinal microbiota are reduced in patients with celiac disease, and on the other hand, pathogenic species are potentially increased compared to healthy people. In these patients, although dysbiosis in the intestinal microbiota is decreased after a GFD, it is not eliminated. Therefore, the intestinal microbiota plays a crucial role in the pathogenesis of the celiac disease [17, 18, 19]. On the other hand, fewer studies have investigated the number and composition of intestinal microbiota and its role in the pathogenesis of the celiac disease, as well as comparing the composition of intestinal microbiota in people with celiac disease compared to people without this disease. Therefore, in the current study, the composition of specific intestinal microbiota, including Bifidobacterium and Lactobacillus, was investigated in patients with celiac disease compared to healthy individuals, and it was shown that the population of intestinal microbiota in celiac patients is significantly different compared to healthy individuals so that sick people have a lower amount of beneficial intestinal bacteria Bifidobacterium and Lactobacillus than healthy people. The diversity of intestinal microbiota in people with celiac disease compared to healthy people has been investigated in various studies that confirm the results of the present study [10, 20]. Golfetto et al. agreed with the results of the present study regarding the low level of Bifidobacterium and dysbiosis in the intestinal microbiota in patients with celiac disease, even despite following a GFD, which supports the pathological process of the disease [21]. Similar to the present study is the study of Moraes et al. who reported the difference in the microbial profile between children with celiac disease and the control group and showed that celiac patients have a lower amount of Lactobacillus and Bifidobacterium compared to healthy individuals. Studies have shown that Bifidobacterium and Lactobacillus, which are also part of probiotics, can play a role in the digestion or change of gluten polypeptides. On the other hand, some bacterial species belonging to the genera Lactobacillus and Bifidobacterium have a protective role on epithelial cells against the damage caused by gliadin [22]. Collado et al. in a study identified specific intestinal bacteria related to celiac disease in the diagnosis and post-treatment with GFD in stool samples of children with untreated celiac disease and under GFD. Biopsy samples of untreated celiac patients under GFD and stool samples and biopsies of healthy children as a control group were examined to compare the two groups and real-time PCR was used to measure intestinal bacteria. The results of this study show the difference between the number of some intestinal microbiota, such as Bacteroidetes and Clostridium leptum in children with celiac disease compared to the control group regardless of the stage of the disease, as well as the difference in the number of E.coli. and showed Staphylococcus in untreated celiac children compared to the control group. Also, in this study, a lower amount of Bifidobacterium was reported in the feces of both groups of patients and biopsies of untreated individuals compared to the control group [10]. In another similar study, Sanz et al. analyzed the fecal microbiota count in the population of children with celiac disease compared to age-matched controls by a polymerase chain reaction and denaturing gradient gel electrophoresis and reported a significant increase in the stool microbiota population in patients with celiac disease compared to healthy individuals. On the other hand, they observed the specific presence of Lactobacillus curvatus species in celiac patients and Lactobacillus casei as a characteristic bacterial species in the healthy group. Also, the number of Bifidobacterium species in the celiac group was significantly lower than in the healthy group [21]. Nistal et al. conducted a study to investigate the difference in intestinal microbiota in adults with celiac disease and healthy individuals. Using the techniques of gradient genetic electrophoresis and gas-liquid chromatography of short-chain fatty acids, the microbial communities were measured in stool samples of untreated celiac patients, celiac patients treated with GFD, and healthy population and they observed a decrease in the diversity of Lactobacillus and Bifidobacterium species in treated celiac patients. Also, the treated celiac group showed a significantly higher amount of Bifidobacterium bifidum than healthy adults. The overall results of this study showed the difference in the fecal microbiota of untreated celiac patients compared to healthy people. Consistent with the present study, Nistal et al. showed that although the GFD in celiac patients partially restores the digestive microbiota to a normal state, the diversity of Lactobacillus and Bifidobacterium decreases significantly [23]. In another similar study conducted by Nylund et al. on patients with celiac disease and gluten-sensitive people without celiac disease under a GFD and healthy people consuming oats, the composition of fecal microbiota in the studied groups was investigated and the results showed that although the frequency of Bifidobacterium in healthy adults tended to be higher compared to celiac patients and the non-celiac gluten-sensitive group, it did not show a significant difference in terms of diversity in the microbial composition in the studied groups [33]. Also, Di Biase et al. compared the composition of the digestive microbiota in the stool samples of twelve children with celiac disease at the beginning of the disease with the healthy group. The results of this study are also similar to the current study, the difference in the digestive microbiota in children showed celiac disease at the beginning of the disease compared to the healthy group. A decrease in the abundance of beneficial bacteria, such as Bacteroides/Prevotella and Akkermansia was observed in stool samples of celiac disease patients compared to the healthy group [34]. In addition, some other studies also confirm the results of the present study and prove that two beneficial bacteria, Bifidobacterium and Lactobacillus, can play a protective role in patients with celiac disease against the inflammatory response and mucosal damage caused by gliadin peptides. They also explain the therapeutic roles of these probiotics [18, 19, 20, 21, 22, 23, 24]. Reducing the number of beneficial bacteria, such as Lactobacillus and Bifidobacterium in patients with celiac disease leads to an increase in opportunistic pathogens in patients with celiac disease and eventually leads to a defect in the immune system of these patients [26]. According to previous studies, the intestinal microbiota in patients with celiac disease is not completely restored despite following a GFD [10] and the amount of some useful digestive bacteria, such as Bifidobacterium in people with celiac disease is still significantly lower than in healthy people despite this type of diet. The results of most studies conducted in the field of investigating the diversity of the intestinal microbiota of celiac patients under a GFD and comparing it with healthy people all over the world show the diversity and a different number of the intestinal microbiota of these patients and healthy people. [27]. All these results confirm the results of the present study. In the current study, the target population was middle-aged patients and healthy people in an urban society with an average and normal diet. Compared to the group of healthy people, these people had an imbalance in the number of natural probiotics of flora. The amount of Lactobacillus and Bifidobacterium excreted in these people is less than in the healthy group, which is a reflection of the condition of the intestinal flora of these people, which is caused by the poor condition of the microbial flora and the lack of probiotic bacteria in the digestive system of these people. On the other hand, some studies showed no difference between the intestinal microbiota of patients with the celiac disease under a GFD and the control group and declared that the intestinal microbiota does not seem to play a role in the pathogenesis of the celiac disease [31]. On the other hand, having this type of diet in these patients, based on the results of previous studies, only leads to the improvement of a part of the intestinal microbiota [32], but the reasons are not known. However, factors such as the genetics of a patient with celiac disease, despite a GFD, can most likely affect the composition of the intestinal microbiota [33]. On the other hand, gluten has a prebiotic-like function, and removing this protein from a GFD can also create a different intestinal microbiota in these patients compared to healthy people [34]. Therefore, it can be said that modifying the nutritional system with the approach of strengthening the probiotic system of the digestive system and regulating their reliability and viability can moderate the complications of celiac disease, which is a kind of defect in the gene system with epigenetic stimuli. Physiologically, according to the atrophy of the intestinal villi in this disease, perhaps the defect in maintaining the balance of the natural microbial flora can be considered as one of the complications caused by this disease, in another approach, this factor can be considered as the cause of the exacerbation of this disease. According to other effective environmental and biological factors, this series of factors is the occurrence of nutritional poverty and deficiencies in the natural levels of micronutrients. Factors in the present study, the intestinal microbiota and the decrease in the amount of Bifidobacterium and Lactobacillus were different between the two studied groups and decreased in the group with celiac disease, but according to the sample size reported, this difference between the microbiota cannot be determined with certainty. Rohedei reported that the studies on healthy and celiac patients need to have a larger sample size. As shown in the findings section, the results of two Pearson chi-square tests for categorical variables and t test for numerical variables, which were performed to determine the integrity between the two groups of cases and controls in the statistical analysis of demographic data, show that the two studied groups have no significant differences in terms of demographic data. This problem is the strength of the present study in the sense that these results have confirmed the integration between the two studied groups and the two groups can be easily compared and examined. The present study had limitations. First, this study was conducted in a small sample size and a single center in Tehran City, Iran. Another limitation of the present study was the limited duration of the study and patient selection.
Conclusion
The present study, consistent with other studies, shows the phenomenon of imbalance of intestinal microbiota in patients with celiac disease compared to healthy people. As a result, it can be said that examining stool microbiota can be a good indicator of intestinal microbiota imbalance in celiac patients and can be used to monitor microbiota restoration during a gluten-free diet. The metabolic data along with the assessment of the intestinal microbiota of patients can help doctors evaluate the role of intestinal microbiota in the pathogenesis of celiac disease and also further investigations.
Ethical Considerations
Compliance with ethical guidelines
Following the principles of research ethics, the researchers followed all ethical codes related to research on human samples and obtained the necessary permits from the competent authorities from the Ethics Committee of the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences and Health Services (Ethical Code: IR.SBMU.RIGLD.REC.1395.114). Also, registration code IRCT20171225038063N1 of the Iran Clinical Trial Registration Center (IRCT) was obtained.
Funding
This research received financial support from the Digestive and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences and Health Services.
Authors' contributions
All authors equally contributed to preparing this article.
Conflicts of interest
The authors declared no conflict of interest.
Acknowledgements
The authors of this article are grateful for the unwavering support of the vice president of research and the colleagues of the Research Institute of Gastrointestinal and Liver Diseases of Shahid Beheshti University of Medical Sciences and the respected officials of the Islamic Azad University, Karaj branch.
References
- Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. Journal of Medical Microbiology. 2007; 56(Pt 12):1669-74. [DOI:10.1099/jmm.0.47410-0] [PMID]
- Di Sabatino A, Corazza GR. Coeliac disease. Lancet. 2009; 373(9673):1480-93. [DOI:10.1016/S0140-6736(09)60254-3] [PMID]
- Kaukinen K, Lindfors K, Collin P, Koskinen O, Mäki M. Coeliac disease-a diagnostic and therapeutic challenge. Clinical Chemistry and Laboratory Medicine. 2010; 48(9):1205-16. [DOI:10.1515/cclm.2010.241] [PMID]
- Catassi C, Kryszak D, Bhatti B, Sturgeon C, Helzlsouer K, Clipp SL, et al. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Annals of Medicine. 2010; 42(7):530-8. [DOI:10.3109/07853890.2010.514285] [PMID]
- Sanz Y, De Pama G, Laparra M. Unraveling the ties between celiac disease and intestinal microbiota. International Reviews of Immunology. 2011; 30(4):207-18. [DOI:10.3109/08830185.2011.599084] [PMID]
- Ghosh S. Advances in our understanding of the pathogenesis of celiac disease. Canadian Journal of Gastroenterology . 2011; 25(4):186. [DOI:10.1155/2011/684230] [PMID] [PMCID]
- Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003; 361(9356):512-9. [DOI:10.1016/S0140-6736(03)12489-0] [PMID]
- Cristofori F, Indrio F, Miniello VL, De Angelis M, Francavilla R. Probiotics in celiac disease. Nutrients. 2018; 10(12):1824. [DOI:10.3390/nu10121824] [PMID] [PMCID]
- Nakayama J, Kobayashi T, Tanaka S, Korenori Y, Tateyama A, Sakamoto N, et al. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunology and Medical Microbiology. 2011; 63(3):397-406. [DOI:10.1111/j.1574-695X.2011.00872.x] [PMID]
- Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. Journal of Clinical Pathology. 2009; 62(3):264-9. [DOI:10.1136/jcp.2008.061366] [PMID]
- Davis CD, Milner JA. Gastrointestinal microflora, food components and colon cancer prevention. The Journal of Nutritional Biochemistry. 2009; 20(10):743-52. [DOI:10.1016/j.jnutbio.2009.06.001] [PMID] [PMCID]
- Ou G, Hedberg M, Horstedt P, Baranov V, Forsberg G, Drobni M,et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. The American Journal of Gastroenterology. 2009; 104(12):3058-67. [DOI:10.1038/ajg.2009.524] [PMID]
- Sanders ME. Impact of probiotics on colonizing microbiota of the gut. Journal of Clinical Gastroenterology. 2011; 45(S):S115-9. [DOI:10.1097/MCG.0b013e318227414a] [PMID]
- Marsh MN, Johnson MW, Rostami K. Mucosal histopathology in celiac disease: A rebuttal of Oberhuber’s sub-division of Marsh III. Gastroenterology and Hepatology from Bed to Bench. 2015; 8(2):99-109. [PMID]
- Chen H, Rangasamy M, Tan SY, Wang H, Siegfried BD. Evaluation of five methods for total DNA extraction from western corn rootworm beetles. PLoS One. 2010; 5(8):e11963. [DOI:10.1371/journal.pone.0011963] [PMID] [PMCID]
- Wang IK, Lai HC, Yu CJ, Liang CC, Chang CT, Kuo HL, et al. Real-time PCR analysis of the intestinal microbiotas in peritoneal dialysis patients. Applied and environmental microbiology. 2012; 78(4):1107-12. [DOI:10.1128/AEM.05605-11] [PMID] [PMCID]
- Marasco G, Di Biase AR, Schiumerini R, Eusebi LH, Iughetti L, Ravaioli F, et al. Gut microbiota and celiac disease. Digestive Diseases and Sciences. 2016; 61(6):1461-72. [DOI:10.1007/s10620-015-4020-2] [PMID]
- Aureli P, Capurso L, Castellazzi AM, Clerici M, Giovannini M, Morelli L, et al. Probiotics and health: An evidence-based review. Pharmacological Research. 2011; 63(5):366-76. [DOI:10.1016/j.phrs.2011.02.006] [PMID]
- Harnett J, Myers SP, Rolfe M. Probiotics and the microbiome in celiac disease: A randomised controlled trial. Evidence-Based Complementary and Alternative Medicine. 2016; 2016:9048574. [DOI:10.1155/2016/9048574] [PMID] [PMCID]
- Grover S, Rashmi HM, Srivastava AK, Batish VK. Probiotics for human health -new innovations and emerging trends. Gut pathogens. 2012; 4(1):15. [DOI:10.1186/1757-4749-4-15] [PMID] [PMCID
- Sanz Y, Sanchez E, Marzotto M, Calabuig M, Torriani S, Dellaglio F. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunology and Medical Microbiology. 2007; 51(3):562-8. [DOI:10.1111/j.1574-695X.2007.00337.x] [PMID]
- Golfetto L, Duarte de Senna F, Hermes J, Beserra BTS, Franca F, Martinello F. Lower bifidobacteria counts in adult patients with celiac disease on a gluten-free diet. Arquivos de Gastroenterologia. 2014; 51(2):139-43. [DOI:10.1590/S0004-28032014000200013] [PMID]
- Moraes LF, Grzeskowiak LM, Teixeira TF, Gouveia Peluzio M. Intestinal microbiota and probiotics in celiac disease. Clinical Microbiology Reviews. 2014; 27(3):482-9. [DOI:10.1128/CMR.00106-13] [PMID] [PMCID]
- Nistal E, Caminero A, Vivas S, Ruiz de Morales JM, Saenz de Miera LE, Rodriguez-Aparicio LB, et al. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie. 2012; 94(8):1724-9. [DOI:10.1016/j.biochi.2012.03.025] [PMID]
- Abdullah GA, Jamalludeen NM, Mansour AA. The role of probiotics in celiac disease and their potential effect on immunological and clinical markers of the disease. International Journal of Scientific & Engineering Research. 2017; 8(9):1347-73. [Link]
- Di Biase AR, Marasco G, Ravaioli F, Dajti E, Colecchia L, Righi B, et al. Gut microbiota signatures and clinical manifestations in celiac disease children at onset: A pilot study. Journal of Gastroenterology and Hepatology. 2021; 36(2):446-54. [DOI:10.1111/jgh.15183] [PMID]
- Sanchez E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal Staphylococcus spp. and virulent features associated with coeliac disease. Journal of Clinical Pathology. 2012; 65(9):830-4. [DOI:10.1136/jclinpath-2012-200759] [PMID]
- Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiology. 2008; 8:232. [DOI:10.1186/1471-2180-8-232] [PMID] [PMCID]
- De Meij TG, Budding AE, Grasman ME, Kneepkens CM, Savelkoul PH, Mearin ML. Composition and diversity of the duodenal mucosa-associated microbiome in children with untreated coeliac disease. Scandinavian Journal of Gastroenterology. 2013; 48(5):530-6. [DOI:10.3109/00365521.2013.775666] [PMID]
- Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, et al. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiology. 2010; 10:175. [DOI:10.1186/1471-2180-10-175] [PMID] [PMCID]
- Olivares M, Neef A, Castillejo G, Palma GD, Varea V, Capilla A, et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2015; 64(3):406-17. [DOI:10.1136/gutjnl-2014-306931] [PMID]
- De Palma G, Nadal I, Collado MC, Sanz Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. The British Journal of Nutrition. 2009; 102(8):1154-60. [DOI:10.1017/S0007114509371767] [PMID]
- Nylund L, Hakkola S, Lahti L, Salminen S, Kalliomäki M, Yang B, et al. Diet, perceived intestinal well-being and compositions of fecal microbiota and short chain fatty acids in oat-using subjects with celiac disease or gluten sensitivity. Nutrients. 2020; 12(9):2570.[DOI:10.3390/nu12092570] [PMID] [PMCID]
- Chander AM, Yadav H, Jain S, Bhadada SK, Dhawan DK. Cross-talk between gluten, intestinal microbiota and intestinal mucosa in Celiac disease: Recent advances and basis of autoimmunity. Frontiers in Microbiology. 2018; 9:2597. [DOI:10.3389/fmicb.2018.02597] [PMID] [PMCID]
Type of Study: Original |
Subject:
Basic Medical Science
Received: 2021/12/27 | Accepted: 2022/06/22 | Published: 2022/07/1
Received: 2021/12/27 | Accepted: 2022/06/22 | Published: 2022/07/1
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |